Selective Suppression of the Splicing-Mediated MicroRNA Pathway by the Terminal Uridyltransferase Tailor
- PMID: 26145174
- PMCID: PMC4517475
- DOI: 10.1016/j.molcel.2015.05.034
Selective Suppression of the Splicing-Mediated MicroRNA Pathway by the Terminal Uridyltransferase Tailor
Abstract
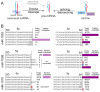
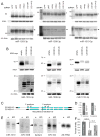
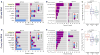
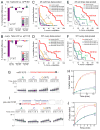
Several terminal uridyltransferases (TUTases) are known to modulate small RNA biogenesis and/or function via diverse mechanisms. Here, we demonstrate that Drosophila splicing-derived pre-miRNAs (mirtrons) are efficiently modified by the previously uncharacterized TUTase, Tailor. Tailor is necessary and sufficient for mirtron hairpin uridylation, and this modification inhibits mirtron biogenesis. Genome-wide analyses demonstrate that mirtrons are dominant Tailor substrates, and three features contribute to substrate specificity. First, reprogramming experiments show Tailor preferentially identifies splicing-derived miRNAs. Second, in vitro tests indicate Tailor prefers substrate hairpins over mature miRNAs. Third, Tailor exhibits sequence preference for 3'-terminal AG, a defining mirtron characteristic. Our work supports the notion that Tailor preferentially suppresses biogenesis of mirtrons, an evolutionarily adventitious pre-miRNA substrate class. Moreover, we detect preferential activity of Tailor on 3'-G canonical pre-miRNAs, and specific depletion of such loci from the pool of conserved miRNAs. Thus, Tailor activity may have had collateral impact on shaping populations of canonical miRNAs.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Characterization of a TUTase/RNase complex required for Drosophila gametogenesis.RNA. 2017 Mar;23(3):284-296. doi: 10.1261/rna.059527.116. Epub 2016 Dec 14. RNA. 2017. PMID: 27974621 Free PMC article.
-
Uridylation of RNA Hairpins by Tailor Confines the Emergence of MicroRNAs in Drosophila.Mol Cell. 2015 Jul 16;59(2):203-16. doi: 10.1016/j.molcel.2015.05.033. Epub 2015 Jul 2. Mol Cell. 2015. PMID: 26145176 Free PMC article.
-
Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.Nucleic Acids Res. 2019 Jan 10;47(1):495-508. doi: 10.1093/nar/gky1116. Nucleic Acids Res. 2019. PMID: 30407553 Free PMC article.
-
Mirtrons: microRNA biogenesis via splicing.Biochimie. 2011 Nov;93(11):1897-904. doi: 10.1016/j.biochi.2011.06.017. Epub 2011 Jun 21. Biochimie. 2011. PMID: 21712066 Free PMC article. Review.
-
Biogenesis, characterization, and functions of mirtrons.Wiley Interdiscip Rev RNA. 2022 Jan;13(1):e1680. doi: 10.1002/wrna.1680. Epub 2021 Jun 21. Wiley Interdiscip Rev RNA. 2022. PMID: 34155810 Review.
Cited by
-
Promiscuous splicing-derived hairpins are dominant substrates of tailing-mediated defense of miRNA biogenesis in mammals.Cell Rep. 2023 Feb 28;42(2):112111. doi: 10.1016/j.celrep.2023.112111. Epub 2023 Feb 16. Cell Rep. 2023. PMID: 36800291 Free PMC article.
-
Characterization of a TUTase/RNase complex required for Drosophila gametogenesis.RNA. 2017 Mar;23(3):284-296. doi: 10.1261/rna.059527.116. Epub 2016 Dec 14. RNA. 2017. PMID: 27974621 Free PMC article.
-
Non-canonical features of microRNAs: paradigms emerging from cardiovascular disease.Nat Rev Cardiol. 2022 Sep;19(9):620-638. doi: 10.1038/s41569-022-00680-2. Epub 2022 Mar 18. Nat Rev Cardiol. 2022. PMID: 35304600 Review.
-
AGO-bound mature miRNAs are oligouridylated by TUTs and subsequently degraded by DIS3L2.Nat Commun. 2020 Jun 2;11(1):2765. doi: 10.1038/s41467-020-16533-w. Nat Commun. 2020. PMID: 32488030 Free PMC article.
-
Characterisation of the in-vivo miRNA landscape in Drosophila ribonuclease mutants reveals Pacman-mediated regulation of the highly conserved let-7 cluster during apoptotic processes.Front Genet. 2024 Feb 20;15:1272689. doi: 10.3389/fgene.2024.1272689. eCollection 2024. Front Genet. 2024. PMID: 38444757 Free PMC article.
References
-
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nature genetics. 2004;5:396–400. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

