Double-Stranded RNA Is Detected by Immunofluorescence Analysis in RNA and DNA Virus Infections, Including Those by Negative-Stranded RNA Viruses
- PMID: 26136565
- PMCID: PMC4542381
- DOI: 10.1128/JVI.01299-15
Double-Stranded RNA Is Detected by Immunofluorescence Analysis in RNA and DNA Virus Infections, Including Those by Negative-Stranded RNA Viruses
Abstract
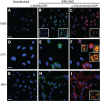
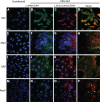
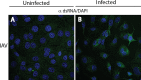
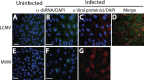
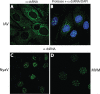
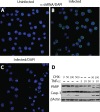
Early biochemical studies of viral replication suggested that most viruses produce double-stranded RNA (dsRNA), which is essential for the induction of the host immune response. However, it was reported in 2006 that dsRNA could be detected by immunofluorescence antibody staining in double-stranded DNA and positive-strand RNA virus infections but not in negative-strand RNA virus infections. Other reports in the literature seemed to support these observations. This suggested that negative-strand RNA viruses produce little, if any, dsRNA or that more efficient viral countermeasures to mask dsRNA are mounted. Because of our interest in the use of dsRNA antibodies for virus discovery, particularly in pathological specimens, we wanted to determine how universal immunostaining for dsRNA might be in animal virus infections. We have detected the in situ formation of dsRNA in cells infected with vesicular stomatitis virus, measles virus, influenza A virus, and Nyamanini virus, which represent viruses from different negative-strand RNA virus families. dsRNA was also detected in cells infected with lymphocytic choriomeningitis virus, an ambisense RNA virus, and minute virus of mice (MVM), a single-stranded DNA (ssDNA) parvovirus, but not hepatitis B virus. Although dsRNA staining was primarily observed in the cytoplasm, it was also seen in the nucleus of cells infected with influenza A virus, Nyamanini virus, and MVM. Thus, it is likely that most animal virus infections produce dsRNA species that can be detected by immunofluorescence staining. The apoptosis induced in several uninfected cell lines failed to upregulate dsRNA formation.
Importance: An effective antiviral host immune response depends on recognition of viral invasion and an intact innate immune system as a first line of defense. Double-stranded RNA (dsRNA) is a viral product essential for the induction of innate immunity, leading to the production of type I interferons (IFNs) and the activation of hundreds of IFN-stimulated genes. The present study demonstrates that infections, including those by ssDNA viruses and positive- and negative-strand RNA viruses, produce dsRNAs detectable by standard immunofluorescence staining. While dsRNA staining was primarily observed in the cytoplasm, nuclear staining was also present in some RNA and DNA virus infections. The nucleus is unlikely to have pathogen-associated molecular pattern (PAMP) receptors for dsRNA because of the presence of host dsRNA molecules. Thus, it is likely that most animal virus infections produce dsRNA species detectable by immunofluorescence staining, which may prove useful in viral discovery as well.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses.J Virol. 2006 May;80(10):5059-64. doi: 10.1128/JVI.80.10.5059-5064.2006. J Virol. 2006. PMID: 16641297 Free PMC article.
-
Confocal Imaging of Double-Stranded RNA and Pattern Recognition Receptors in Negative-Sense RNA Virus Infection.J Vis Exp. 2019 Jan 26;(143):10.3791/59095. doi: 10.3791/59095. J Vis Exp. 2019. PMID: 30741258 Free PMC article.
-
Genome-Wide CRISPR-Cas9 Screen Reveals the Importance of the Heparan Sulfate Pathway and the Conserved Oligomeric Golgi Complex for Synthetic Double-Stranded RNA Uptake and Sindbis Virus Infection.mSphere. 2020 Nov 11;5(6):e00914-20. doi: 10.1128/mSphere.00914-20. mSphere. 2020. PMID: 33177215 Free PMC article.
-
Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit.Antiviral Res. 2013 Dec;100(3):615-35. doi: 10.1016/j.antiviral.2013.10.002. Epub 2013 Oct 12. Antiviral Res. 2013. PMID: 24129118 Free PMC article. Review.
-
A Novel Mechanism Underlying the Innate Immune Response Induction upon Viral-Dependent Replication of Host Cell mRNA: A Mistake of +sRNA Viruses' Replicases.Front Cell Infect Microbiol. 2017 Jan 20;7:5. doi: 10.3389/fcimb.2017.00005. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28164038 Free PMC article. Review.
Cited by
-
ADAR2 Is Involved in Self and Nonself Recognition of Borna Disease Virus Genomic RNA in the Nucleus.J Virol. 2020 Feb 28;94(6):e01513-19. doi: 10.1128/JVI.01513-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31852792 Free PMC article.
-
Puumala and Andes Orthohantaviruses Cause Transient Protein Kinase R-Dependent Formation of Stress Granules.J Virol. 2020 Jan 17;94(3):e01168-19. doi: 10.1128/JVI.01168-19. Print 2020 Jan 17. J Virol. 2020. PMID: 31723021 Free PMC article.
-
Next-generation sequencing of dsRNA is greatly improved by treatment with the inexpensive denaturing reagent DMSO.Microb Genom. 2019 Nov;5(11):e000315. doi: 10.1099/mgen.0.000315. Microb Genom. 2019. PMID: 31738702 Free PMC article.
-
Influenza Virus Infection Induces ZBP1 Expression and Necroptosis in Mouse Lungs.Front Cell Infect Microbiol. 2019 Aug 7;9:286. doi: 10.3389/fcimb.2019.00286. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31440477 Free PMC article.
-
Tunneling nanotubes provide a route for SARS-CoV-2 spreading.Sci Adv. 2022 Jul 22;8(29):eabo0171. doi: 10.1126/sciadv.abo0171. Epub 2022 Jul 20. Sci Adv. 2022. PMID: 35857849 Free PMC article.
References
-
- Williams BR. 2001. Signal integration via PKR. Sci STKE 2001:re2. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

