FUS functions in coupling transcription to splicing by mediating an interaction between RNAP II and U1 snRNP
- PMID: 26124092
- PMCID: PMC4507187
- DOI: 10.1073/pnas.1506282112
FUS functions in coupling transcription to splicing by mediating an interaction between RNAP II and U1 snRNP
Abstract
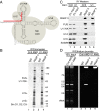
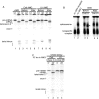
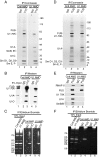
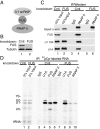
Pre-mRNA splicing is coupled to transcription by RNA polymerase II (RNAP II). We previously showed that U1 small nuclear ribonucleoprotein (snRNP) associates with RNAP II, and both RNAP II and U1 snRNP are also the most abundant factors associated with the protein fused-in-sarcoma (FUS), which is mutated to cause the neurodegenerative disease amyotrophic lateral sclerosis. Here, we show that an antisense morpholino that base-pairs to the 5' end of U1 snRNA blocks splicing in the coupled system and completely disrupts the association between U1 snRNP and both FUS and RNAP II, but has no effect on the association between FUS and RNAP II. Conversely, we found that U1 snRNP does not interact with RNAP II in FUS knockdown extracts. Moreover, using these extracts, we found that FUS must be present during the transcription reaction in order for splicing to occur. Together, our data lead to a model that FUS functions in coupling transcription to splicing via mediating an interaction between RNAP II and U1 snRNP.
Keywords: ALS; RNA polymerase II; U1 snRNP; coupling transcription to splicing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
SR proteins function in coupling RNAP II transcription to pre-mRNA splicing.Mol Cell. 2007 Jun 22;26(6):867-81. doi: 10.1016/j.molcel.2007.05.036. Mol Cell. 2007. PMID: 17588520
-
Interactome analyses revealed that the U1 snRNP machinery overlaps extensively with the RNAP II machinery and contains multiple ALS/SMA-causative proteins.Sci Rep. 2018 Jun 8;8(1):8755. doi: 10.1038/s41598-018-27136-3. Sci Rep. 2018. PMID: 29884807 Free PMC article.
-
Splicing-independent recruitment of U1 snRNP to a transcription unit in living cells.J Cell Sci. 2010 Jun 15;123(Pt 12):2085-93. doi: 10.1242/jcs.061358. J Cell Sci. 2010. PMID: 20519584
-
[The role of the U1 snRNP complex in the regulation of gene expression: recent reports].Postepy Biochem. 2024 Sep 17;70(3):348-357. doi: 10.18388/pb.2021_566. Print 2024 Sep 30. Postepy Biochem. 2024. PMID: 39365565 Review. Polish.
-
U1 snRNP Biogenesis Defects in Neurodegenerative Diseases.Chembiochem. 2024 May 2;25(9):e202300864. doi: 10.1002/cbic.202300864. Epub 2024 Mar 25. Chembiochem. 2024. PMID: 38459794 Review.
Cited by
-
An autoregulation loop in fust-1 for circular RNA regulation in Caenorhabditis elegans.Genetics. 2021 Nov 5;219(3):iyab145. doi: 10.1093/genetics/iyab145. Genetics. 2021. PMID: 34740247 Free PMC article.
-
FUS-dependent microRNA deregulations identify TRIB2 as a druggable target for ALS motor neurons.iScience. 2023 Oct 6;26(11):108152. doi: 10.1016/j.isci.2023.108152. eCollection 2023 Nov 17. iScience. 2023. PMID: 37920668 Free PMC article.
-
The hnRNP RALY regulates transcription and cell proliferation by modulating the expression of specific factors including the proliferation marker E2F1.J Biol Chem. 2017 Dec 1;292(48):19674-19692. doi: 10.1074/jbc.M117.795591. Epub 2017 Sep 27. J Biol Chem. 2017. PMID: 28972179 Free PMC article.
-
FUS interacts with nuclear matrix-associated protein SAFB1 as well as Matrin3 to regulate splicing and ligand-mediated transcription.Sci Rep. 2016 Oct 12;6:35195. doi: 10.1038/srep35195. Sci Rep. 2016. PMID: 27731383 Free PMC article.
-
LINC01106 drives colorectal cancer growth and stemness through a positive feedback loop to regulate the Gli family factors.Cell Death Dis. 2020 Oct 16;11(10):869. doi: 10.1038/s41419-020-03026-3. Cell Death Dis. 2020. PMID: 33067422 Free PMC article.
References
-
- Greenleaf AL. Positive patches and negative noodles: Linking RNA processing to transcription? Trends Biochem Sci. 1993;18(4):117–119. - PubMed
-
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416(6880):499–506. - PubMed
-
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: An answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21(1):11–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

