Structure-based design of potent HIV-1 protease inhibitors with modified P1-biphenyl ligands: synthesis, biological evaluation, and enzyme-inhibitor X-ray structural studies
- PMID: 26107245
- PMCID: PMC4765733
- DOI: 10.1021/acs.jmedchem.5b00676
Structure-based design of potent HIV-1 protease inhibitors with modified P1-biphenyl ligands: synthesis, biological evaluation, and enzyme-inhibitor X-ray structural studies
Abstract
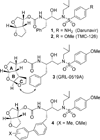
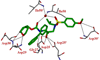
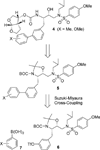
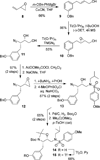
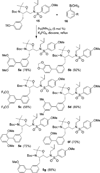
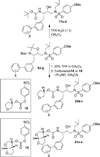
We report the design, synthesis, X-ray structural studies, and biological evaluation of a novel series of HIV-1 protease inhibitors. We designed a variety of functionalized biphenyl derivatives to make enhanced van der Waals interactions in the S1 subsite of HIV-1 protease. These biphenyl derivatives were conveniently synthesized using a Suzuki-Miyaura cross-coupling reaction as the key step. We examined the potential of these functionalized biphenyl-derived P1 ligands in combination with 3-(S)-tetrahydrofuranyl urethane and bis-tetrahydrofuranyl urethane as the P2 ligands. Inhibitor 21e, with a 2-methoxy-1,1'-biphenyl derivative as P1 ligand and bis-THF as the P2 ligand, displayed the most potent enzyme inhibitory and antiviral activity. This inhibitor also exhibited potent activity against a panel of multidrug-resistant HIV-1 variants. A high resolution X-ray crystal structure of related Boc-derivative 17a-bound HIV-1 protease provided important molecular insight into the ligand-binding site interactions of the biphenyl core in the S1 subsite of HIV-1 protease.
Figures

Similar articles
-
Design of HIV-1 protease inhibitors with C3-substituted hexahydrocyclopentafuranyl urethanes as P2-ligands: synthesis, biological evaluation, and protein-ligand X-ray crystal structure.J Med Chem. 2011 Aug 25;54(16):5890-901. doi: 10.1021/jm200649p. Epub 2011 Jul 29. J Med Chem. 2011. PMID: 21800876 Free PMC article.
-
Design and Synthesis of Highly Potent HIV-1 Protease Inhibitors Containing Tricyclic Fused Ring Systems as Novel P2 Ligands: Structure-Activity Studies, Biological and X-ray Structural Analysis.J Med Chem. 2018 May 24;61(10):4561-4577. doi: 10.1021/acs.jmedchem.8b00298. Epub 2018 May 15. J Med Chem. 2018. PMID: 29763303 Free PMC article.
-
Design and synthesis of potent HIV-1 protease inhibitors incorporating hexahydrofuropyranol-derived high affinity P(2) ligands: structure-activity studies and biological evaluation.J Med Chem. 2011 Jan 27;54(2):622-34. doi: 10.1021/jm1012787. Epub 2010 Dec 31. J Med Chem. 2011. PMID: 21194227 Free PMC article.
-
Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance.Acc Chem Res. 2008 Jan;41(1):78-86. doi: 10.1021/ar7001232. Epub 2007 Aug 28. Acc Chem Res. 2008. PMID: 17722874 Review.
-
Darunavir, a conceptually new HIV-1 protease inhibitor for the treatment of drug-resistant HIV.Bioorg Med Chem. 2007 Dec 15;15(24):7576-80. doi: 10.1016/j.bmc.2007.09.010. Epub 2007 Sep 14. Bioorg Med Chem. 2007. PMID: 17900913 Free PMC article. Review.
Cited by
-
Design, Synthesis, and Biological Evaluation of Darunavir Analogs as HIV-1 Protease Inhibitors.ACS Bio Med Chem Au. 2024 Sep 19;4(5):242-256. doi: 10.1021/acsbiomedchemau.4c00040. eCollection 2024 Oct 16. ACS Bio Med Chem Au. 2024. PMID: 39431267 Free PMC article.
-
Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.J Med Chem. 2016 Jul 28;59(14):6826-37. doi: 10.1021/acs.jmedchem.6b00639. Epub 2016 Jul 7. J Med Chem. 2016. PMID: 27389367 Free PMC article.
-
Diastereoselective Synthesis of the HIV Protease Inhibitor Darunavir and Related Derivatives via a Titanium Tetrachloride-Mediated Asymmetric Glycolate Aldol Addition Reaction.J Org Chem. 2024 Jul 5;89(13):9569-9585. doi: 10.1021/acs.joc.4c01057. Epub 2024 Jun 25. J Org Chem. 2024. PMID: 38916048 Free PMC article.
-
Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS.J Med Chem. 2016 Jun 9;59(11):5172-208. doi: 10.1021/acs.jmedchem.5b01697. Epub 2016 Jan 22. J Med Chem. 2016. PMID: 26799988 Free PMC article. Review.
-
An NMR strategy to detect conformational differences in a protein complexed with highly analogous inhibitors in solution.Methods. 2018 Sep 15;148:9-18. doi: 10.1016/j.ymeth.2018.04.005. Epub 2018 Apr 12. Methods. 2018. PMID: 29656080 Free PMC article.
References
-
- Mitsuya H, Maeda K, Das D, Ghosh AK. Development of protease inhibitors and the fight with drug-resistant HIV-1 variants. Adv. Pharmacol. 2008;56:169–197. - PubMed
-
- Conway B. HAART in Treatment--experienced Patients in the 21st Century: The Audacity of Hope. Future Virol. 2009;4:39–41.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Miscellaneous

