Alternatively Spliced Homologous Exons Have Ancient Origins and Are Highly Expressed at the Protein Level
- PMID: 26061177
- PMCID: PMC4465641
- DOI: 10.1371/journal.pcbi.1004325
Alternatively Spliced Homologous Exons Have Ancient Origins and Are Highly Expressed at the Protein Level
Abstract
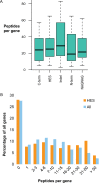
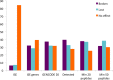
Alternative splicing of messenger RNA can generate a wide variety of mature RNA transcripts, and these transcripts may produce protein isoforms with diverse cellular functions. While there is much supporting evidence for the expression of alternative transcripts, the same is not true for the alternatively spliced protein products. Large-scale mass spectroscopy experiments have identified evidence of alternative splicing at the protein level, but with conflicting results. Here we carried out a rigorous analysis of the peptide evidence from eight large-scale proteomics experiments to assess the scale of alternative splicing that is detectable by high-resolution mass spectroscopy. We find fewer splice events than would be expected: we identified peptides for almost 64% of human protein coding genes, but detected just 282 splice events. This data suggests that most genes have a single dominant isoform at the protein level. Many of the alternative isoforms that we could identify were only subtly different from the main splice isoform. Very few of the splice events identified at the protein level disrupted functional domains, in stark contrast to the two thirds of splice events annotated in the human genome that would lead to the loss or damage of functional domains. The most striking result was that more than 20% of the splice isoforms we identified were generated by substituting one homologous exon for another. This is significantly more than would be expected from the frequency of these events in the genome. These homologous exon substitution events were remarkably conserved--all the homologous exons we identified evolved over 460 million years ago--and eight of the fourteen tissue-specific splice isoforms we identified were generated from homologous exons. The combination of proteomics evidence, ancient origin and tissue-specific splicing indicates that isoforms generated from homologous exons may have important cellular roles.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Comparative proteomics reveals a significant bias toward alternative protein isoforms with conserved structure and function.Mol Biol Evol. 2012 Sep;29(9):2265-83. doi: 10.1093/molbev/mss100. Epub 2012 Mar 22. Mol Biol Evol. 2012. PMID: 22446687 Free PMC article.
-
The evolutionary fate of alternatively spliced homologous exons after gene duplication.Genome Biol Evol. 2015 Apr 29;7(6):1392-403. doi: 10.1093/gbe/evv076. Genome Biol Evol. 2015. PMID: 25931610 Free PMC article.
-
An analysis of tissue-specific alternative splicing at the protein level.PLoS Comput Biol. 2020 Oct 5;16(10):e1008287. doi: 10.1371/journal.pcbi.1008287. eCollection 2020 Oct. PLoS Comput Biol. 2020. PMID: 33017396 Free PMC article.
-
Alternative Splicing May Not Be the Key to Proteome Complexity.Trends Biochem Sci. 2017 Feb;42(2):98-110. doi: 10.1016/j.tibs.2016.08.008. Epub 2016 Oct 3. Trends Biochem Sci. 2017. PMID: 27712956 Free PMC article. Review.
-
Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology.Cell. 2000 Oct 27;103(3):367-70. doi: 10.1016/s0092-8674(00)00128-8. Cell. 2000. PMID: 11081623 Review. No abstract available.
Cited by
-
Mutant mice lacking alternatively spliced p53 isoforms unveil Ackr4 as a male-specific prognostic factor in Myc-driven B-cell lymphomas.Elife. 2024 Sep 19;13:RP92774. doi: 10.7554/eLife.92774. Elife. 2024. PMID: 39298333 Free PMC article.
-
SAPFIR: A webserver for the identification of alternative protein features.BMC Bioinformatics. 2022 Jun 24;23(1):250. doi: 10.1186/s12859-022-04804-w. BMC Bioinformatics. 2022. PMID: 35751026 Free PMC article.
-
Bridging the Gap Between Environmental Adversity and Neuropsychiatric Disorders: The Role of Transposable Elements.Front Genet. 2022 May 25;13:813510. doi: 10.3389/fgene.2022.813510. eCollection 2022. Front Genet. 2022. PMID: 35711940 Free PMC article. Review.
-
Human canonical CD157/Bst1 is an alternatively spliced isoform masking a previously unidentified primate-specific exon included in a novel transcript.Sci Rep. 2017 Nov 21;7(1):15923. doi: 10.1038/s41598-017-16184-w. Sci Rep. 2017. PMID: 29162908 Free PMC article.
-
The ribosome-engaged landscape of alternative splicing.Nat Struct Mol Biol. 2016 Dec;23(12):1117-1123. doi: 10.1038/nsmb.3317. Epub 2016 Nov 7. Nat Struct Mol Biol. 2016. PMID: 27820807 Free PMC article.
References
-
- Smith CW, Valcárcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25: 381–388. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

