Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1
- PMID: 26056265
- PMCID: PMC4485101
- DOI: 10.1073/pnas.1509744112
Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1
Abstract
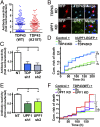
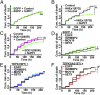
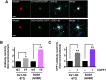
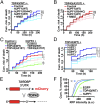
Over 30% of patients with amyotrophic lateral sclerosis (ALS) exhibit cognitive deficits indicative of frontotemporal dementia (FTD), suggesting a common pathogenesis for both diseases. Consistent with this hypothesis, neuronal and glial inclusions rich in TDP43, an essential RNA-binding protein, are found in the majority of those with ALS and FTD, and mutations in TDP43 and a related RNA-binding protein, FUS, cause familial ALS and FTD. TDP43 and FUS affect the splicing of thousands of transcripts, in some cases triggering nonsense-mediated mRNA decay (NMD), a highly conserved RNA degradation pathway. Here, we take advantage of a faithful primary neuronal model of ALS and FTD to investigate and characterize the role of human up-frameshift protein 1 (hUPF1), an RNA helicase and master regulator of NMD, in these disorders. We show that hUPF1 significantly protects mammalian neurons from both TDP43- and FUS-related toxicity. Expression of hUPF2, another essential component of NMD, also improves survival, whereas inhibiting NMD prevents rescue by hUPF1, suggesting that hUPF1 acts through NMD to enhance survival. These studies emphasize the importance of RNA metabolism in ALS and FTD, and identify a uniquely effective therapeutic strategy for these disorders.
Keywords: ALS; FTD; RNA binding proteins; RNA decay; neurodegeneration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay.Proc Natl Acad Sci U S A. 2018 Dec 18;115(51):E11904-E11913. doi: 10.1073/pnas.1810413115. Epub 2018 Nov 19. Proc Natl Acad Sci U S A. 2018. PMID: 30455313 Free PMC article.
-
Complexes between the nonsense-mediated mRNA decay pathway factor human upf1 (up-frameshift protein 1) and essential nonsense-mediated mRNA decay factors in HeLa cells.Biochem J. 2003 Aug 1;373(Pt 3):775-83. doi: 10.1042/BJ20021920. Biochem J. 2003. PMID: 12723973 Free PMC article.
-
Dysfunction in nonsense-mediated decay, protein homeostasis, mitochondrial function, and brain connectivity in ALS-FUS mice with cognitive deficits.Acta Neuropathol Commun. 2021 Jan 6;9(1):9. doi: 10.1186/s40478-020-01111-4. Acta Neuropathol Commun. 2021. PMID: 33407930 Free PMC article.
-
Tale of two diseases: amyotrophic lateral sclerosis and frontotemporal dementia.Neurol India. 2014 Jul-Aug;62(4):347-51. doi: 10.4103/0028-3886.141174. Neurol India. 2014. PMID: 25237937 Review.
-
[Overlapping features of frontotemporal dementia and amyotrophic lateral sclerosis].Rev Med Chil. 2014 Jul;142(7):867-79. doi: 10.4067/S0034-98872014000700007. Rev Med Chil. 2014. PMID: 25378006 Review. Spanish.
Cited by
-
Inhibition of Nonsense-Mediated Decay Induces Nociceptive Sensitization through Activation of the Integrated Stress Response.J Neurosci. 2023 Apr 19;43(16):2921-2933. doi: 10.1523/JNEUROSCI.1604-22.2023. Epub 2023 Mar 9. J Neurosci. 2023. PMID: 36894318 Free PMC article.
-
Phase separation and pathologic transitions of RNP condensates in neurons: implications for amyotrophic lateral sclerosis, frontotemporal dementia and other neurodegenerative disorders.Front Mol Neurosci. 2023 Sep 1;16:1242925. doi: 10.3389/fnmol.2023.1242925. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37720552 Free PMC article. Review.
-
Translation dysregulation in neurodegenerative disorders.Proc Natl Acad Sci U S A. 2018 Dec 18;115(51):12842-12844. doi: 10.1073/pnas.1818493115. Epub 2018 Nov 30. Proc Natl Acad Sci U S A. 2018. PMID: 30504142 Free PMC article. No abstract available.
-
UPF1 reduces C9orf72 HRE-induced neurotoxicity in the absence of nonsense-mediated decay dysfunction.Cell Rep. 2021 Mar 30;34(13):108925. doi: 10.1016/j.celrep.2021.108925. Cell Rep. 2021. PMID: 33789100 Free PMC article.
-
Identification of a localized nonsense-mediated decay pathway at the endoplasmic reticulum.Genes Dev. 2020 Aug 1;34(15-16):1075-1088. doi: 10.1101/gad.338061.120. Epub 2020 Jul 2. Genes Dev. 2020. PMID: 32616520 Free PMC article.
References
-
- Huisman MHB, et al. Population based epidemiology of amyotrophic lateral sclerosis using capture-recapture methodology. J Neurol Neurosurg Psychiatry. 2011;82(10):1165–1170. - PubMed
-
- Barmada SJ, Finkbeiner S. Pathogenic TARDBP mutations in amyotrophic lateral sclerosis and frontotemporal dementia: Disease-associated pathways. Rev Neurosci. 2010;21(4):251–272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

