Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro): IMPLICATIONS FOR nsp5 REGULATION AND THE DEVELOPMENT OF ANTIVIRALS
- PMID: 26055715
- PMCID: PMC4528106
- DOI: 10.1074/jbc.M115.651463
Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro): IMPLICATIONS FOR nsp5 REGULATION AND THE DEVELOPMENT OF ANTIVIRALS
Abstract
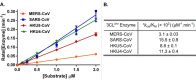
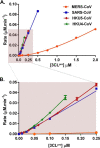
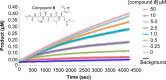
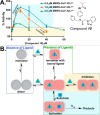
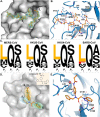
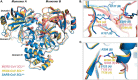
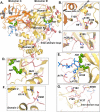
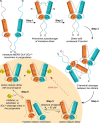
All coronaviruses, including the recently emerged Middle East respiratory syndrome coronavirus (MERS-CoV) from the β-CoV subgroup, require the proteolytic activity of the nsp5 protease (also known as 3C-like protease, 3CL(pro)) during virus replication, making it a high value target for the development of anti-coronavirus therapeutics. Kinetic studies indicate that in contrast to 3CL(pro) from other β-CoV 2c members, including HKU4 and HKU5, MERS-CoV 3CL(pro) is less efficient at processing a peptide substrate due to MERS-CoV 3CL(pro) being a weakly associated dimer. Conversely, HKU4, HKU5, and SARS-CoV 3CL(pro) enzymes are tightly associated dimers. Analytical ultracentrifugation studies support that MERS-CoV 3CL(pro) is a weakly associated dimer (Kd ∼52 μm) with a slow off-rate. Peptidomimetic inhibitors of MERS-CoV 3CL(pro) were synthesized and utilized in analytical ultracentrifugation experiments and demonstrate that MERS-CoV 3CL(pro) undergoes significant ligand-induced dimerization. Kinetic studies also revealed that designed reversible inhibitors act as activators at a low compound concentration as a result of induced dimerization. Primary sequence comparisons and x-ray structural analyses of two MERS-CoV 3CLpro and inhibitor complexes, determined to 1.6 Å, reveal remarkable structural similarity of the dimer interface with 3CL(pro) from HKU4-CoV and HKU5-CoV. Despite this structural similarity, substantial differences in the dimerization ability suggest that long range interactions by the nonconserved amino acids distant from the dimer interface may control MERS-CoV 3CL(pro) dimerization. Activation of MERS-CoV 3CL(pro) through ligand-induced dimerization appears to be unique within the genogroup 2c and may potentially increase the complexity in the development of MERS-CoV 3CL(pro) inhibitors as antiviral agents.
Keywords: MERS-CoV 3CLpro; X-ray crystallography; analytical ultracentrifugation; enzyme inactivation; enzyme inhibitor; enzyme kinetics; ligand-induced dimerization; monomer-dimer equilibrium; viral protease; β-CoV.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Computational modeling of the bat HKU4 coronavirus 3CLpro inhibitors as a tool for the development of antivirals against the emerging Middle East respiratory syndrome (MERS) coronavirus.J Mol Recognit. 2017 Nov;30(11):e2644. doi: 10.1002/jmr.2644. Epub 2017 Jun 13. J Mol Recognit. 2017. PMID: 28608547 Free PMC article.
-
Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4--The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS).Bioorg Med Chem. 2015 Sep 1;23(17):6036-48. doi: 10.1016/j.bmc.2015.06.039. Epub 2015 Jun 19. Bioorg Med Chem. 2015. PMID: 26190463 Free PMC article.
-
Structural-based virtual screening and in vitro assays for small molecules inhibiting the feline coronavirus 3CL protease as a surrogate platform for coronaviruses.Antiviral Res. 2020 Oct;182:104927. doi: 10.1016/j.antiviral.2020.104927. Epub 2020 Sep 7. Antiviral Res. 2020. PMID: 32910955 Free PMC article.
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
-
The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds.Antiviral Res. 2015 Mar;115:21-38. doi: 10.1016/j.antiviral.2014.12.015. Epub 2014 Dec 29. Antiviral Res. 2015. PMID: 25554382 Free PMC article. Review.
Cited by
-
The recent outbreaks of human coronaviruses: A medicinal chemistry perspective.Med Res Rev. 2021 Jan;41(1):72-135. doi: 10.1002/med.21724. Epub 2020 Aug 27. Med Res Rev. 2021. PMID: 32852058 Free PMC article. Review.
-
Druggable targets from coronaviruses for designing new antiviral drugs.Bioorg Med Chem. 2020 Nov 15;28(22):115745. doi: 10.1016/j.bmc.2020.115745. Epub 2020 Sep 8. Bioorg Med Chem. 2020. PMID: 33007557 Free PMC article. Review.
-
The in-vitro effect of famotidine on sars-cov-2 proteases and virus replication.Sci Rep. 2021 Mar 8;11(1):5433. doi: 10.1038/s41598-021-84782-w. Sci Rep. 2021. PMID: 33686143 Free PMC article.
-
The Nsp12-coding region of type 2 PRRSV is required for viral subgenomic mRNA synthesis.Emerg Microbes Infect. 2019;8(1):1501-1510. doi: 10.1080/22221751.2019.1679010. Emerg Microbes Infect. 2019. PMID: 31631782 Free PMC article.
-
Predicted coronavirus Nsp5 protease cleavage sites in the human proteome.BMC Genom Data. 2022 Apr 4;23(1):25. doi: 10.1186/s12863-022-01044-y. BMC Genom Data. 2022. PMID: 35379171 Free PMC article.
References
-
- Barber D. M., Nettleton P. F., Herring J. A. (1985) Disease in a dairy herd associated with the introduction and spread of bovine virus diarrhoea virus. Vet. Rec. 117, 459–464 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

