Crystal structure of TRIM20 C-terminal coiled-coil/B30.2 fragment: implications for the recognition of higher order oligomers
- PMID: 26043233
- PMCID: PMC4455283
- DOI: 10.1038/srep10819
Crystal structure of TRIM20 C-terminal coiled-coil/B30.2 fragment: implications for the recognition of higher order oligomers
Abstract
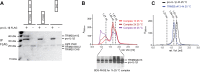
Many tripartite motif-containing (TRIM) proteins, comprising RING-finger, B-Box, and coiled-coil domains, carry additional B30.2 domains on the C-terminus of the TRIM motif and are considered to be pattern recognition receptors involved in the detection of higher order oligomers (e.g. viral capsid proteins). To investigate the spatial architecture of domains in TRIM proteins we determined the crystal structure of the TRIM20Δ413 fragment at 2.4 Å resolution. This structure comprises the central helical scaffold (CHS) and C-terminal B30.2 domains and reveals an anti-parallel arrangement of CHS domains placing the B-box domains 170 Å apart from each other. Small-angle X-ray scattering confirmed that the linker between CHS and B30.2 domains is flexible in solution. The crystal structure suggests an interaction between the B30.2 domain and an extended stretch in the CHS domain, which involves residues that are mutated in the inherited disease Familial Mediterranean Fever. Dimerization of B30.2 domains by means of the CHS domain is crucial for TRIM20 to bind pro-IL-1β in vitro. To exemplify how TRIM proteins could be involved in binding higher order oligomers we discuss three possible models for the TRIM5α/HIV-1 capsid interaction assuming different conformations of B30.2 domains.
Figures



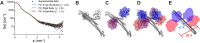

Similar articles
-
The crystal structure of human pyrin b30.2 domain: implications for mutations associated with familial Mediterranean fever.J Mol Biol. 2009 Nov 27;394(2):226-36. doi: 10.1016/j.jmb.2009.08.059. Epub 2009 Aug 31. J Mol Biol. 2009. PMID: 19729025
-
The B30.2 domain of pyrin, the familial Mediterranean fever protein, interacts directly with caspase-1 to modulate IL-1beta production.Proc Natl Acad Sci U S A. 2006 Jun 27;103(26):9982-7. doi: 10.1073/pnas.0602081103. Epub 2006 Jun 19. Proc Natl Acad Sci U S A. 2006. PMID: 16785446 Free PMC article.
-
Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding.Virology. 2006 Sep 15;353(1):234-46. doi: 10.1016/j.virol.2006.05.017. Epub 2006 Jun 30. Virology. 2006. PMID: 16808955
-
Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence?Immunology. 2005 Dec;116(4):411-7. doi: 10.1111/j.1365-2567.2005.02248.x. Immunology. 2005. PMID: 16313355 Free PMC article. Review.
-
The tripartite motif: structure and function.Adv Exp Med Biol. 2012;770:11-25. Adv Exp Med Biol. 2012. PMID: 23630997 Review.
Cited by
-
The Roles of TRIMs in Antiviral Innate Immune Signaling.Front Cell Infect Microbiol. 2021 Mar 15;11:628275. doi: 10.3389/fcimb.2021.628275. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33791238 Free PMC article. Review.
-
Characterisation of assembly and ubiquitylation by the RBCC motif of Trim5α.Sci Rep. 2016 May 27;6:26837. doi: 10.1038/srep26837. Sci Rep. 2016. PMID: 27230667 Free PMC article.
-
Analysis of the Zn-Binding Domains of TRIM32, the E3 Ubiquitin Ligase Mutated in Limb Girdle Muscular Dystrophy 2H.Cells. 2019 Mar 16;8(3):254. doi: 10.3390/cells8030254. Cells. 2019. PMID: 30884854 Free PMC article.
-
TRIM56 coiled-coil domain structure provides insights into its E3 ligase functions.Comput Struct Biotechnol J. 2023 Apr 23;21:2801-2808. doi: 10.1016/j.csbj.2023.04.022. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37168870 Free PMC article.
-
Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5α.Elife. 2016 Jun 2;5:e16309. doi: 10.7554/eLife.16309. Elife. 2016. PMID: 27253059 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

