TP53: an oncogene in disguise
- PMID: 26024390
- PMCID: PMC4495363
- DOI: 10.1038/cdd.2015.53
TP53: an oncogene in disguise
Abstract
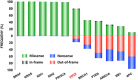
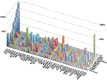
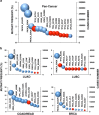
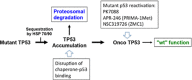
The standard classification used to define the various cancer genes confines tumor protein p53 (TP53) to the role of a tumor suppressor gene. However, it is now an indisputable fact that many p53 mutants act as oncogenic proteins. This statement is based on multiple arguments including the mutation signature of the TP53 gene in human cancer, the various gains-of-function (GOFs) of the different p53 mutants and the heterogeneous phenotypes developed by knock-in mouse strains modeling several human TP53 mutations. In this review, we will shatter the classical and traditional image of tumor protein p53 (TP53) as a tumor suppressor gene by emphasizing its multiple oncogenic properties that make it a potential therapeutic target that should not be underestimated. Analysis of the data generated by the various cancer genome projects highlights the high frequency of TP53 mutations and reveals that several p53 hotspot mutants are the most common oncoprotein variants expressed in several types of tumors. The use of Muller's classical definition of mutations based on quantitative and qualitative consequences on the protein product, such as 'amorph', 'hypomorph', 'hypermorph' 'neomorph' or 'antimorph', allows a more meaningful assessment of the consequences of cancer gene modifications, their potential clinical significance, and clearly demonstrates that the TP53 gene is an atypical cancer gene.
Figures
Similar articles
-
Roles of TP53 in determining therapeutic sensitivity, growth, cellular senescence, invasion and metastasis.Adv Biol Regul. 2017 Jan;63:32-48. doi: 10.1016/j.jbior.2016.10.001. Epub 2016 Oct 6. Adv Biol Regul. 2017. PMID: 27776972 Review.
-
Proteomic identification of heat shock protein 90 as a candidate target for p53 mutation reactivation by PRIMA-1 in breast cancer cells.Breast Cancer Res. 2005;7(5):R765-74. doi: 10.1186/bcr1290. Epub 2005 Jul 27. Breast Cancer Res. 2005. PMID: 16168122 Free PMC article.
-
p53 from basic research to clinical applications.Crit Rev Oncog. 1992;3(3):257-82. Crit Rev Oncog. 1992. PMID: 1616957 Review.
-
High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer.Cancer Res. 2009 Apr 15;69(8):3625-33. doi: 10.1158/0008-5472.CAN-08-3426. Epub 2009 Mar 31. Cancer Res. 2009. PMID: 19336573
-
Mutant p53 accumulation in human breast cancer is not an intrinsic property or dependent on structural or functional disruption but is regulated by exogenous stress and receptor status.J Pathol. 2014 Jul;233(3):238-46. doi: 10.1002/path.4356. Epub 2014 May 21. J Pathol. 2014. PMID: 24687952
Cited by
-
Ultraconserved long non-coding RNA uc.63 in breast cancer.Oncotarget. 2017 May 30;8(22):35669-35680. doi: 10.18632/oncotarget.10572. Oncotarget. 2017. PMID: 27447964 Free PMC article.
-
Cancer and Tumour Suppressor p53 Encounters at the Juncture of Sex Disparity.Front Genet. 2021 Feb 16;12:632719. doi: 10.3389/fgene.2021.632719. eCollection 2021. Front Genet. 2021. PMID: 33664771 Free PMC article. Review.
-
Immunomediated Pan-cancer Regulation Networks are Dominant Fingerprints After Treatment of Cell Lines with Demethylation.Cancer Inform. 2016 Apr 27;15:45-64. doi: 10.4137/CIN.S31809. eCollection 2016. Cancer Inform. 2016. PMID: 27147816 Free PMC article.
-
Combined effect of recombinant human adenovirus p53 and curcumin in the treatment of liver cancer.Exp Ther Med. 2020 Nov;20(5):18. doi: 10.3892/etm.2020.9145. Epub 2020 Aug 26. Exp Ther Med. 2020. PMID: 32934683 Free PMC article.
-
Nkx2.5 Functions as a Conditional Tumor Suppressor Gene in Colorectal Cancer Cells via Acting as a Transcriptional Coactivator in p53-Mediated p21 Expression.Front Oncol. 2021 Apr 1;11:648045. doi: 10.3389/fonc.2021.648045. eCollection 2021. Front Oncol. 2021. PMID: 33869046 Free PMC article.
References
-
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

