Systematic Genome-wide Screening and Prediction of microRNAs in EBOV During the 2014 Ebolavirus Outbreak
- PMID: 26011078
- PMCID: PMC4603304
- DOI: 10.1038/srep09912
Systematic Genome-wide Screening and Prediction of microRNAs in EBOV During the 2014 Ebolavirus Outbreak
Abstract
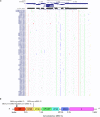
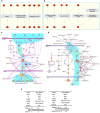
Recently, several thousand people have been killed by the Ebolavirus disease (EVD) in West Africa, yet no current antiviral medications and treatments are available. Systematic investigation of ebolavirus whole genomes during the 2014 outbreak may shed light on the underlying mechanisms of EVD development. Here, using the genome-wide screening in ebolavirus genome sequences, we predicted four putative viral microRNA precursors (pre-miRNAs) and seven putative mature microRNAs (miRNAs). Combing bioinformatics analysis and prediction of the potential ebolavirus miRNA target genes, we suggest that two ebolavirus coding possible miRNAs may be silence and down-regulate the target genes NFKBIE and RIPK1, which are the central mediator of the pathways related with host cell defense mechanism. Additionally, the ebolavirus exploits the miRNAs to inhibit the NF-kB and TNF factors to evade the host defense mechanisms that limit replication by killing infected cells, or to conversely trigger apoptosis as a mechanism to increase virus spreading. This is the first study to use the genome-wide scanning to predict microRNAs in the 2014 outbreak EVD and then to apply systematic bioinformatics to analyze their target genes. We revealed a potential mechanism of miRNAs in ebolavirus infection and possible therapeutic targets for Ebola viral infection treatment.
Figures
Similar articles
-
Identification of Ebola virus microRNAs and their putative pathological function.Sci China Life Sci. 2014 Oct;57(10):973-81. doi: 10.1007/s11427-014-4759-2. Epub 2014 Sep 29. Sci China Life Sci. 2014. PMID: 25266153
-
Ebola virus disease in the Democratic Republic of Congo.N Engl J Med. 2014 Nov 27;371(22):2083-91. doi: 10.1056/NEJMoa1411099. Epub 2014 Oct 15. N Engl J Med. 2014. PMID: 25317743
-
Ebola virus encodes a miR-155 analog to regulate importin-α5 expression.Cell Mol Life Sci. 2016 Oct;73(19):3733-44. doi: 10.1007/s00018-016-2215-0. Epub 2016 Apr 19. Cell Mol Life Sci. 2016. PMID: 27094387 Free PMC article.
-
Treatment of ebola virus disease.Pharmacotherapy. 2015 Jan;35(1):43-53. doi: 10.1002/phar.1545. Pharmacotherapy. 2015. PMID: 25630412 Review.
-
Computational analysis of Ebolavirus data: prospects, promises and challenges.Biochem Soc Trans. 2016 Aug 15;44(4):973-8. doi: 10.1042/BST20160074. Biochem Soc Trans. 2016. PMID: 27528741 Review.
Cited by
-
Computational prediction of miRNAs in Nipah virus genome reveals possible interaction with human genes involved in encephalitis.Mol Biol Res Commun. 2018 Sep;7(3):107-118. doi: 10.22099/mbrc.2018.29577.1322. Mol Biol Res Commun. 2018. PMID: 30426028 Free PMC article.
-
RNA Virus-Encoded miRNAs: Current Insights and Future Challenges.Front Microbiol. 2021 Jun 24;12:679210. doi: 10.3389/fmicb.2021.679210. eCollection 2021. Front Microbiol. 2021. PMID: 34248890 Free PMC article. Review.
-
A Review of the Interaction between miRNAs and Ebola Virus.Int J Mol Cell Med. 2024;13(2):210-219. doi: 10.22088/IJMCM.BUMS.13.2.210. Int J Mol Cell Med. 2024. PMID: 39184819 Free PMC article. Review.
-
Ebola Virus Produces Discrete Small Noncoding RNAs Independently of the Host MicroRNA Pathway Which Lack RNA Interference Activity in Bat and Human Cells.J Virol. 2020 Feb 28;94(6):e01441-19. doi: 10.1128/JVI.01441-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31852785 Free PMC article.
-
Genome-wide computational prediction of miRNAs in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) revealed target genes involved in pulmonary vasculature and antiviral innate immunity.Mol Biol Res Commun. 2020 Jun;9(2):83-91. doi: 10.22099/mbrc.2020.36507.1487. Mol Biol Res Commun. 2020. PMID: 32802902 Free PMC article.
References
-
- Ksiazek T. G. Filoviruses: Marburg and Ebola. Viral Infections of Humans . 14, 337–350 (2014).
-
- Lupkin S., Timeline of the Ebolavirus in America. WABC-TV New York. (2014). Available at: http://7online.com/news/timeline-of-the-ebola-virus-in-america-/348789/. (Accessed 18th October 2014)
-
- World Heath Organization. Ebola response roadmap. (28th Angust 2014)
-
- Frieden T. R., Damon I., Bell B. P., Kenyon T. & Nichol S. Ebola 2014--New Challenges, New Global Response and Responsibility. N. Eng. J. Med. 371, 1117–1180 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

