The nucleoprotein of influenza A virus induces p53 signaling and apoptosis via attenuation of host ubiquitin ligase RNF43
- PMID: 25996295
- PMCID: PMC4669709
- DOI: 10.1038/cddis.2015.131
The nucleoprotein of influenza A virus induces p53 signaling and apoptosis via attenuation of host ubiquitin ligase RNF43
Abstract
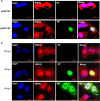
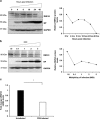
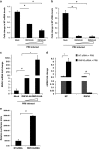
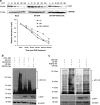
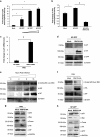
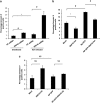
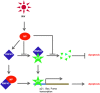
The interplay between influenza virus and host factors to support the viral life cycle is well documented. Influenza A virus (IAV) proteins interact with an array of cellular proteins and hijack host pathways which are at the helm of cellular responses to facilitate virus invasion. The multifaceted nature of the ubiquitination pathway for protein regulation makes it a vulnerable target of many viruses including IAV. To this end we conducted a yeast two-hybrid screen to search for cellular ubiquitin ligases important for influenza virus replication. We identified host protein, RING finger protein 43 (RNF43), a RING-type E3 ubiquitin ligase, as a novel interactor of nucleoprotein (NP) of IAV and an essential partner to induce NP-driven p53-mediated apoptosis in IAV-infected cells. In this study, we demonstrate that IAV leads to attenuation of RNF43 transcripts and hence its respective protein levels in the cellular milieu whereas in RNF43 depleted cells, viral replication was escalated several folds. Moreover, RNF43 polyubiquitinates p53 which further leads to its destabilization resulting in a decrease in induction of the p53 apoptotic pathway, a hitherto unknown process targeted by NP for p53 stabilization and accumulation. Collectively, these results conclude that NP targets RNF43 to modulate p53 ubiquitination levels and hence causes p53 stabilization which is conducive to an enhanced apoptosis level in the host cells. In conclusion, our study unravels a novel strategy adopted by IAV for utilizing the much conserved ubiquitin proteasomal pathway.
Figures

Similar articles
-
TRIM41-Mediated Ubiquitination of Nucleoprotein Limits Influenza A Virus Infection.J Virol. 2018 Jul 31;92(16):e00905-18. doi: 10.1128/JVI.00905-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29899090 Free PMC article.
-
RNF43 interacts with NEDL1 and regulates p53-mediated transcription.Biochem Biophys Res Commun. 2011 Jan 7;404(1):143-7. doi: 10.1016/j.bbrc.2010.11.082. Epub 2010 Nov 23. Biochem Biophys Res Commun. 2011. PMID: 21108931
-
CNOT4-Mediated Ubiquitination of Influenza A Virus Nucleoprotein Promotes Viral RNA Replication.mBio. 2017 May 23;8(3):e00597-17. doi: 10.1128/mBio.00597-17. mBio. 2017. PMID: 28536288 Free PMC article.
-
Essential Roles of E3 Ubiquitin Ligases in p53 Regulation.Int J Mol Sci. 2017 Feb 17;18(2):442. doi: 10.3390/ijms18020442. Int J Mol Sci. 2017. PMID: 28218667 Free PMC article. Review.
-
The Roles of Ubiquitination in Pathogenesis of Influenza Virus Infection.Int J Mol Sci. 2022 Apr 21;23(9):4593. doi: 10.3390/ijms23094593. Int J Mol Sci. 2022. PMID: 35562987 Free PMC article. Review.
Cited by
-
Porcine Circovirus 2 Activates the PERK-Reactive Oxygen Species Axis To Induce p53 Phosphorylation with Subsequent Cell Cycle Arrest at S Phase in Favor of Its Replication.J Virol. 2022 Nov 23;96(22):e0127422. doi: 10.1128/jvi.01274-22. Epub 2022 Oct 27. J Virol. 2022. PMID: 36300938 Free PMC article.
-
Late regulation of immune genes and microRNAs in circulating leukocytes in a pig model of influenza A (H1N2) infection.Sci Rep. 2016 Feb 19;6:21812. doi: 10.1038/srep21812. Sci Rep. 2016. PMID: 26893019 Free PMC article.
-
Nucleocapsid proteins: roles beyond viral RNA packaging.Wiley Interdiscip Rev RNA. 2016 Mar-Apr;7(2):213-26. doi: 10.1002/wrna.1326. Epub 2016 Jan 8. Wiley Interdiscip Rev RNA. 2016. PMID: 26749541 Free PMC article. Review.
-
Comparative Profiling of Ubiquitin Proteasome System Interplay with Influenza A Virus PB2 Polymerase Protein Recapitulating Virus Evolution in Humans.mSphere. 2017 Nov 22;2(6):e00330-17. doi: 10.1128/mSphere.00330-17. eCollection 2017 Nov-Dec. mSphere. 2017. PMID: 29202037 Free PMC article.
-
Abdominal and Pelvic Organ Failure Induced by Intraperitoneal Influenza A Virus Infection in Mice.Front Microbiol. 2020 Jul 17;11:1713. doi: 10.3389/fmicb.2020.01713. eCollection 2020. Front Microbiol. 2020. PMID: 32765481 Free PMC article.
References
-
- Nailwal H, Kamra K, Lal SK. H7N9: a killer in the making or a false alarm? Can J Microbiol 2014; 60: 425–429. - PubMed
-
- Kawaoka Y, Cox NJ, Haller O, Hongo S, Kaverin N, Klenk HD et al. Orthomyxoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds), Virus taxonomy: 8th report of the international committee on taxonomy of viruses. Elsevier Academia Press: London, UK, 2005; pp 681–693.
-
- Portela A, Digard P. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol 2002; 83: 723–734. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

