IMP 2.0: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks
- PMID: 25969450
- PMCID: PMC4489318
- DOI: 10.1093/nar/gkv486
IMP 2.0: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks
Abstract
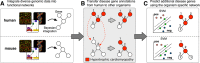
IMP (Integrative Multi-species Prediction), originally released in 2012, is an interactive web server that enables molecular biologists to interpret experimental results and to generate hypotheses in the context of a large cross-organism compendium of functional predictions and networks. The system provides biologists with a framework to analyze their candidate gene sets in the context of functional networks, expanding or refining their sets using functional relationships predicted from integrated high-throughput data. IMP 2.0 integrates updated prior knowledge and data collections from the last three years in the seven supported organisms (Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Danio rerio, Caenorhabditis elegans, and Saccharomyces cerevisiae) and extends function prediction coverage to include human disease. IMP identifies homologs with conserved functional roles for disease knowledge transfer, allowing biologists to analyze disease contexts and predictions across all organisms. Additionally, IMP 2.0 implements a new flexible platform for experts to generate custom hypotheses about biological processes or diseases, making sophisticated data-driven methods easily accessible to researchers. IMP does not require any registration or installation and is freely available for use at http://imp.princeton.edu.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
IMP: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks.Nucleic Acids Res. 2012 Jul;40(Web Server issue):W484-90. doi: 10.1093/nar/gks458. Epub 2012 Jun 7. Nucleic Acids Res. 2012. PMID: 22684505 Free PMC article.
-
FNTM: a server for predicting functional networks of tissues in mouse.Nucleic Acids Res. 2015 Jul 1;43(W1):W182-7. doi: 10.1093/nar/gkv443. Epub 2015 May 4. Nucleic Acids Res. 2015. PMID: 25940632 Free PMC article.
-
GeneMANIA prediction server 2013 update.Nucleic Acids Res. 2013 Jul;41(Web Server issue):W115-22. doi: 10.1093/nar/gkt533. Nucleic Acids Res. 2013. PMID: 23794635 Free PMC article.
-
GeneMANIA update 2018.Nucleic Acids Res. 2018 Jul 2;46(W1):W60-W64. doi: 10.1093/nar/gky311. Nucleic Acids Res. 2018. PMID: 29912392 Free PMC article.
-
The Alliance of Genome Resources: Building a Modern Data Ecosystem for Model Organism Databases.Genetics. 2019 Dec;213(4):1189-1196. doi: 10.1534/genetics.119.302523. Genetics. 2019. PMID: 31796553 Free PMC article. Review.
Cited by
-
Induced Torpor as a Countermeasure for Low Dose Radiation Exposure in a Zebrafish Model.Cells. 2021 Apr 14;10(4):906. doi: 10.3390/cells10040906. Cells. 2021. PMID: 33920039 Free PMC article.
-
Applications of comparative evolution to human disease genetics.Curr Opin Genet Dev. 2015 Dec;35:16-24. doi: 10.1016/j.gde.2015.08.004. Epub 2015 Sep 4. Curr Opin Genet Dev. 2015. PMID: 26338499 Free PMC article. Review.
-
Machine Learning Analysis Identifies Drosophila Grunge/Atrophin as an Important Learning and Memory Gene Required for Memory Retention and Social Learning.G3 (Bethesda). 2017 Nov 6;7(11):3705-3718. doi: 10.1534/g3.117.300172. G3 (Bethesda). 2017. PMID: 28889104 Free PMC article.
-
Computational strategies for cross-species knowledge transfer and translational biomedicine.ArXiv [Preprint]. 2024 Aug 16:arXiv:2408.08503v1. ArXiv. 2024. PMID: 39184546 Free PMC article. Preprint.
-
Genome-wide functional association networks: background, data & state-of-the-art resources.Brief Bioinform. 2020 Jul 15;21(4):1224-1237. doi: 10.1093/bib/bbz064. Brief Bioinform. 2020. PMID: 31281921 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

