The Phyre2 web portal for protein modeling, prediction and analysis
- PMID: 25950237
- PMCID: PMC5298202
- DOI: 10.1038/nprot.2015.053
The Phyre2 web portal for protein modeling, prediction and analysis
Abstract
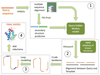
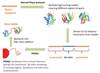
Phyre2 is a suite of tools available on the web to predict and analyze protein structure, function and mutations. The focus of Phyre2 is to provide biologists with a simple and intuitive interface to state-of-the-art protein bioinformatics tools. Phyre2 replaces Phyre, the original version of the server for which we previously published a paper in Nature Protocols. In this updated protocol, we describe Phyre2, which uses advanced remote homology detection methods to build 3D models, predict ligand binding sites and analyze the effect of amino acid variants (e.g., nonsynonymous SNPs (nsSNPs)) for a user's protein sequence. Users are guided through results by a simple interface at a level of detail they determine. This protocol will guide users from submitting a protein sequence to interpreting the secondary and tertiary structure of their models, their domain composition and model quality. A range of additional available tools is described to find a protein structure in a genome, to submit large number of sequences at once and to automatically run weekly searches for proteins that are difficult to model. The server is available at http://www.sbg.bio.ic.ac.uk/phyre2. A typical structure prediction will be returned between 30 min and 2 h after submission.
Conflict of interest statement
MJES is a Director and shareholder in Equinox Pharma Ltd which uses bioinformatics and chemoinformatics in drug discovery research and services.
Figures




Similar articles
-
Protein structure prediction on the Web: a case study using the Phyre server.Nat Protoc. 2009;4(3):363-71. doi: 10.1038/nprot.2009.2. Nat Protoc. 2009. PMID: 19247286
-
IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences.Nucleic Acids Res. 2015 Jul 1;43(W1):W169-73. doi: 10.1093/nar/gkv236. Epub 2015 Mar 27. Nucleic Acids Res. 2015. PMID: 25820431 Free PMC article.
-
3DLigandSite: predicting ligand-binding sites using similar structures.Nucleic Acids Res. 2010 Jul;38(Web Server issue):W469-73. doi: 10.1093/nar/gkq406. Epub 2010 May 31. Nucleic Acids Res. 2010. PMID: 20513649 Free PMC article.
-
State-of-the-art bioinformatics protein structure prediction tools (Review).Int J Mol Med. 2011 Sep;28(3):295-310. doi: 10.3892/ijmm.2011.705. Epub 2011 May 23. Int J Mol Med. 2011. PMID: 21617841 Review.
-
Computational tools for protein modeling.Curr Protein Pept Sci. 2000 Jul;1(1):1-21. doi: 10.2174/1389203003381469. Curr Protein Pept Sci. 2000. PMID: 12369918 Review.
Cited by
-
Decoding the genomic terrain: functional insights into 14 chemosensory proteins in whitefly Bemisia tabaci Asia II-1.Sci Rep. 2024 Nov 1;14(1):26252. doi: 10.1038/s41598-024-77998-z. Sci Rep. 2024. PMID: 39482332
-
Identification of potential bioactive phytochemicals for the inhibition of platelet-derived growth factor receptor β: a structure-based approach for cancer therapy.Front Mol Biosci. 2024 Oct 15;11:1492847. doi: 10.3389/fmolb.2024.1492847. eCollection 2024. Front Mol Biosci. 2024. PMID: 39473823 Free PMC article.
-
Identification of a novel Golgi-localized putative glycosyltransferase protein in Arabidopsis thaliana.Plant Biotechnol (Tokyo). 2024 Mar 25;41(1):35-44. doi: 10.5511/plantbiotechnology.23.1214a. Plant Biotechnol (Tokyo). 2024. PMID: 39464868 Free PMC article.
-
Reexamining the Mycovirome of Botrytis spp.Viruses. 2024 Oct 21;16(10):1640. doi: 10.3390/v16101640. Viruses. 2024. PMID: 39459972 Free PMC article.
-
Identification of a Novel Antiviral Lectin against SARS-CoV-2 Omicron Variant from Shiitake-Mushroom-Derived Vesicle-like Nanoparticles.Viruses. 2024 Sep 30;16(10):1546. doi: 10.3390/v16101546. Viruses. 2024. PMID: 39459880 Free PMC article.
References
-
- Mukherjee Srayanta, Szilagyi Andras, Roy Ambrish, Zhang Yang. Genome-wide protein structure prediction. Multiscale approaches to protein modeling: structure prediction, dynamics, thermodynamics and macromolecular assemblies. In: Kolinski Andrzej., editor. Chapter 11. Springer; London: 2010. pp. 255–280.
-
- Koonin EV, et al. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. - PubMed
-
- Kelley LA, Sternberg MJE. Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. - PubMed
-
- Mao C, et al. Functional assignment of Mycobacterium tuberculosis proteome by genome-scale fold-recognition. Tuberculosis. 2013;1:93. - PubMed
Publication types
MeSH terms
Grants and funding
- G1000390-1/1/MRC_/Medical Research Council/United Kingdom
- BB/G022569/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F020481/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G003912/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J019240/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

