Transgenic systems for unequivocal identification of cardiac myocyte nuclei and analysis of cardiomyocyte cell cycle status
- PMID: 25925989
- PMCID: PMC4414935
- DOI: 10.1007/s00395-015-0489-2
Transgenic systems for unequivocal identification of cardiac myocyte nuclei and analysis of cardiomyocyte cell cycle status
Abstract
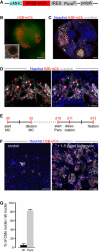
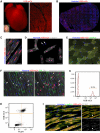
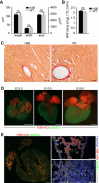
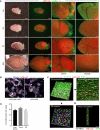
Even though the mammalian heart has been investigated for many years, there are still uncertainties in the fields of cardiac cell biology and regeneration with regard to exact fractions of cardiomyocytes (CMs) at different developmental stages, their plasticity after cardiac lesion and also their basal turnover rate. A main shortcoming is the accurate identification of CM and the demonstration of CM division. Therefore, an in vivo model taking advantage of a live reporter-based identification of CM nuclei and their cell cycle status is needed. In this technical report, we describe the generation and characterization of embryonic stem cells and transgenic mice expressing a fusion protein of human histone 2B and the red fluorescence protein mCherry under control of the CM specific αMHC promoter. This fluorescence label allows unequivocal identification and quantitation of CM nuclei and nuclearity in isolated cells and native tissue slices. In ventricles of adults, we determined a fraction of <20 % CMs and binucleation of 77-90 %, while in atria a CM fraction of 30 % and a binucleation index of 14 % were found. We combined this transgenic system with the CAG-eGFP-anillin transgene, which identifies cell division and established a novel screening assay for cell cycle-modifying substances in isolated, postnatal CMs. Our transgenic live reporter-based system enables reliable identification of CM nuclei and determination of CM fractions and nuclearity in heart tissue. In combination with CAG-eGFP-anillin-mice, the cell cycle status of CMs can be monitored in detail enabling screening for proliferation-inducing substances in vitro and in vivo.
Figures

Similar articles
-
Midbody Positioning and Distance Between Daughter Nuclei Enable Unequivocal Identification of Cardiomyocyte Cell Division in Mice.Circ Res. 2018 Oct 12;123(9):1039-1052. doi: 10.1161/CIRCRESAHA.118.312792. Circ Res. 2018. PMID: 30355161
-
Visualization of Cell Cycle Variations and Determination of Nucleation in Postnatal Cardiomyocytes.J Vis Exp. 2017 Feb 24;(120):55204. doi: 10.3791/55204. J Vis Exp. 2017. PMID: 28287537 Free PMC article.
-
Time-lapse imaging of cell cycle dynamics during development in living cardiomyocyte.J Mol Cell Cardiol. 2014 Jul;72:241-9. doi: 10.1016/j.yjmcc.2014.03.020. Epub 2014 Apr 3. J Mol Cell Cardiol. 2014. PMID: 24704900
-
Can the cardiomyocyte cell cycle be reprogrammed?J Mol Cell Cardiol. 2007 Apr;42(4):706-21. doi: 10.1016/j.yjmcc.2007.01.006. Epub 2007 Jan 24. J Mol Cell Cardiol. 2007. PMID: 17362983 Review.
-
Heart regeneration and the cardiomyocyte cell cycle.Pflugers Arch. 2018 Feb;470(2):241-248. doi: 10.1007/s00424-017-2061-4. Epub 2017 Aug 28. Pflugers Arch. 2018. PMID: 28849267 Free PMC article. Review.
Cited by
-
Cardiomyocyte renewal in the human heart: insights from the fall-out.Eur Heart J. 2017 Aug 7;38(30):2333-2342. doi: 10.1093/eurheartj/ehx343. Eur Heart J. 2017. PMID: 28810672 Free PMC article. Review.
-
Neonatal Growth Restriction Slows Cardiomyocyte Development and Reduces Adult Heart Size.Anat Rec (Hoboken). 2018 Aug;301(8):1398-1404. doi: 10.1002/ar.23851. Epub 2018 May 20. Anat Rec (Hoboken). 2018. PMID: 29729218 Free PMC article.
-
Transient pacing in pigs with complete heart block via myocardial injection of mRNA coding for the T-box transcription factor 18.Nat Biomed Eng. 2024 Sep;8(9):1124-1141. doi: 10.1038/s41551-024-01211-9. Epub 2024 May 2. Nat Biomed Eng. 2024. PMID: 38698155 Free PMC article.
-
Role of Mononuclear Cardiomyocytes in Cardiac Turnover and Regeneration.Curr Cardiol Rep. 2020 May 19;22(6):39. doi: 10.1007/s11886-020-01289-y. Curr Cardiol Rep. 2020. PMID: 32430578 Free PMC article. Review.
-
A protein tertiary structure mimetic modulator of the Hippo signalling pathway.Nat Commun. 2020 Oct 27;11(1):5425. doi: 10.1038/s41467-020-19224-8. Nat Commun. 2020. PMID: 33110077 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

