The Endoplasmic Reticulum Stress Sensor IRE1α in Intestinal Epithelial Cells Is Essential for Protecting against Colitis
- PMID: 25925952
- PMCID: PMC4463471
- DOI: 10.1074/jbc.M114.633560
The Endoplasmic Reticulum Stress Sensor IRE1α in Intestinal Epithelial Cells Is Essential for Protecting against Colitis
Abstract
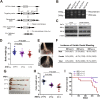
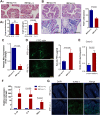
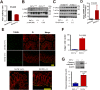
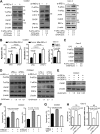
Intestinal epithelial cells (IECs) have critical roles in maintaining homeostasis of intestinal epithelium. Endoplasmic reticulum (ER) stress is implicated in intestinal epithelium homeostasis and inflammatory bowel disease; however, it remains elusive whether IRE1α, a major sensor of ER stress, is directly involved in these processes. We demonstrate here that genetic ablation of Ire1α in IECs leads to spontaneous colitis in mice. Deletion of IRE1α in IECs results in loss of goblet cells and failure of intestinal epithelial barrier function. IRE1α deficiency induces cell apoptosis through induction of CHOP, the pro-apoptotic protein, and sensitizes cells to lipopolysaccharide, an endotoxin from bacteria. IRE1α deficiency confers upon mice higher susceptibility to chemical-induced colitis. These results suggest that IRE1α functions to maintain the intestinal epithelial homeostasis and plays an important role in defending against inflammation bowel diseases.
Keywords: colitis; endoplasmic reticulum stress (ER stress); inflammation; inflammatory bowel disease (IBD); intestinal epithelium.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
Similar articles
-
Activation of pH-Sensing Receptor OGR1 (GPR68) Induces ER Stress Via the IRE1α/JNK Pathway in an Intestinal Epithelial Cell Model.Sci Rep. 2020 Jan 29;10(1):1438. doi: 10.1038/s41598-020-57657-9. Sci Rep. 2020. PMID: 31996710 Free PMC article.
-
Fortilin binds IRE1α and prevents ER stress from signaling apoptotic cell death.Nat Commun. 2017 May 26;8(1):18. doi: 10.1038/s41467-017-00029-1. Nat Commun. 2017. PMID: 28550308 Free PMC article.
-
IRE1α is an endogenous substrate of endoplasmic-reticulum-associated degradation.Nat Cell Biol. 2015 Dec;17(12):1546-55. doi: 10.1038/ncb3266. Epub 2015 Nov 9. Nat Cell Biol. 2015. PMID: 26551274 Free PMC article.
-
IRE1α Implications in Endoplasmic Reticulum Stress-Mediated Development and Pathogenesis of Autoimmune Diseases.Front Immunol. 2018 Jun 6;9:1289. doi: 10.3389/fimmu.2018.01289. eCollection 2018. Front Immunol. 2018. PMID: 29928282 Free PMC article. Review.
-
The unfolded protein response: integrating stress signals through the stress sensor IRE1α.Physiol Rev. 2011 Oct;91(4):1219-43. doi: 10.1152/physrev.00001.2011. Physiol Rev. 2011. PMID: 22013210 Review.
Cited by
-
Sirtuin 1 alleviates endoplasmic reticulum stress-mediated apoptosis of intestinal epithelial cells in ulcerative colitis.World J Gastroenterol. 2019 Oct 14;25(38):5800-5813. doi: 10.3748/wjg.v25.i38.5800. World J Gastroenterol. 2019. PMID: 31636473 Free PMC article.
-
Endoplasmic Reticulum Stress in Colonic Mucosa of Ulcerative Colitis Patients Is Mediated by PERK and IRE1 Pathway Activation.Mediators Inflamm. 2022 Feb 9;2022:6049500. doi: 10.1155/2022/6049500. eCollection 2022. Mediators Inflamm. 2022. PMID: 35185383 Free PMC article.
-
Elevation in Cell Cycle and Protein Metabolism Gene Transcription in Inactive Colonic Tissue From Icelandic Patients With Ulcerative Colitis.Inflamm Bowel Dis. 2019 Jan 10;25(2):317-327. doi: 10.1093/ibd/izy350. Inflamm Bowel Dis. 2019. PMID: 30452647 Free PMC article.
-
Amino Acids in Endoplasmic Reticulum Stress and Redox Signaling.Adv Exp Med Biol. 2021;1332:35-49. doi: 10.1007/978-3-030-74180-8_3. Adv Exp Med Biol. 2021. PMID: 34251637
-
FKBP11 protects intestinal epithelial cells against inflammation‑induced apoptosis via the JNK‑caspase pathway in Crohn's disease.Mol Med Rep. 2018 Nov;18(5):4428-4438. doi: 10.3892/mmr.2018.9485. Epub 2018 Sep 14. Mol Med Rep. 2018. PMID: 30221722 Free PMC article.
References
-
- Baumgart D. C., Carding S. R. (2007) Inflammatory bowel disease: cause and immunobiology. Lancet 369, 1627–1640 - PubMed
-
- Rutgeerts P., Vermeire S., Van Assche G. (2009) Biological therapies for inflammatory bowel diseases. Gastroenterology 136, 1182–1197 - PubMed
-
- Hall P. A., Coates P. J., Ansari B., Hopwood D. (1994) Regulation of cell number in the mammalian gastrointestinal tract: the importance of apoptosis. J. Cell Sci. 107, 3569–3577 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

