Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing
- PMID: 25910207
- PMCID: PMC4410947
- DOI: 10.1016/j.cell.2015.03.027
Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing
Abstract
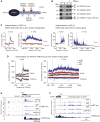
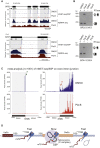
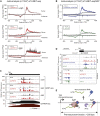
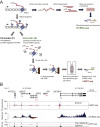
Transcription is a highly dynamic process. Consequently, we have developed native elongating transcript sequencing technology for mammalian chromatin (mNET-seq), which generates single-nucleotide resolution, nascent transcription profiles. Nascent RNA was detected in the active site of RNA polymerase II (Pol II) along with associated RNA processing intermediates. In particular, we detected 5'splice site cleavage by the spliceosome, showing that cleaved upstream exon transcripts are associated with Pol II CTD phosphorylated on the serine 5 position (S5P), which is accumulated over downstream exons. Also, depletion of termination factors substantially reduces Pol II pausing at gene ends, leading to termination defects. Notably, termination factors play an additional promoter role by restricting non-productive RNA synthesis in a Pol II CTD S2P-specific manner. Our results suggest that CTD phosphorylation patterns established for yeast transcription are significantly different in mammals. Taken together, mNET-seq provides dynamic and detailed snapshots of the complex events underlying transcription in mammals.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





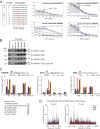
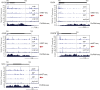



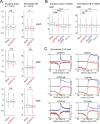
Similar articles
-
Mammalian NET-seq analysis defines nascent RNA profiles and associated RNA processing genome-wide.Nat Protoc. 2016 Mar;11(3):413-28. doi: 10.1038/nprot.2016.012. Epub 2016 Feb 4. Nat Protoc. 2016. PMID: 26844429 Free PMC article.
-
Analysis of Mammalian Native Elongating Transcript sequencing (mNET-seq) high-throughput data.Methods. 2020 Jun 1;178:89-95. doi: 10.1016/j.ymeth.2019.09.003. Epub 2019 Sep 5. Methods. 2020. PMID: 31493517
-
RNA Polymerase II Phosphorylated on CTD Serine 5 Interacts with the Spliceosome during Co-transcriptional Splicing.Mol Cell. 2018 Oct 18;72(2):369-379.e4. doi: 10.1016/j.molcel.2018.09.004. Mol Cell. 2018. PMID: 30340024 Free PMC article.
-
Use of the nuclear walk-on methodology to determine sites of RNA polymerase II initiation and pausing and quantify nascent RNAs in cells.Methods. 2019 Apr 15;159-160:165-176. doi: 10.1016/j.ymeth.2019.02.003. Epub 2019 Feb 8. Methods. 2019. PMID: 30743000 Free PMC article. Review.
-
Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription.Biochim Biophys Acta. 2013 Jan;1829(1):55-62. doi: 10.1016/j.bbagrm.2012.08.013. Epub 2012 Sep 7. Biochim Biophys Acta. 2013. PMID: 22982363 Review.
Cited by
-
Systematic perturbations of SETD2, NSD1, NSD2, NSD3, and ASH1L reveal their distinct contributions to H3K36 methylation.Genome Biol. 2024 Oct 10;25(1):263. doi: 10.1186/s13059-024-03415-3. Genome Biol. 2024. PMID: 39390582 Free PMC article.
-
Lights, camera, action! Capturing the spliceosome and pre-mRNA splicing with single-molecule fluorescence microscopy.Wiley Interdiscip Rev RNA. 2016 Sep;7(5):683-701. doi: 10.1002/wrna.1358. Epub 2016 May 20. Wiley Interdiscip Rev RNA. 2016. PMID: 27198613 Free PMC article. Review.
-
New means to an end: mRNA export activity impacts alternative polyadenylation.Transcription. 2019 Aug-Oct;10(4-5):207-211. doi: 10.1080/21541264.2019.1658557. Epub 2019 Sep 2. Transcription. 2019. PMID: 31474181 Free PMC article.
-
Nascent RNA analyses: tracking transcription and its regulation.Nat Rev Genet. 2019 Dec;20(12):705-723. doi: 10.1038/s41576-019-0159-6. Epub 2019 Aug 9. Nat Rev Genet. 2019. PMID: 31399713 Free PMC article. Review.
-
Lessons from eRNAs: understanding transcriptional regulation through the lens of nascent RNAs.Transcription. 2020 Feb;11(1):3-18. doi: 10.1080/21541264.2019.1704128. Epub 2019 Dec 19. Transcription. 2020. PMID: 31856658 Free PMC article. Review.
References
-
- Adelman K., Marr M.T., Werner J., Saunders A., Ni Z., Andrulis E.D., Lis J.T. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell. 2005;17:103–112. - PubMed
-
- Ahn S.H., Kim M., Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. - PubMed
Supplemental References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

