Systematic interaction network filtering identifies CRMP1 as a novel suppressor of huntingtin misfolding and neurotoxicity
- PMID: 25908449
- PMCID: PMC4417118
- DOI: 10.1101/gr.182444.114
Systematic interaction network filtering identifies CRMP1 as a novel suppressor of huntingtin misfolding and neurotoxicity
Abstract
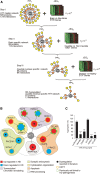
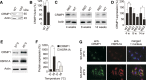
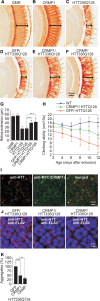
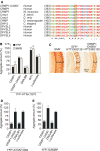
Assemblies of huntingtin (HTT) fragments with expanded polyglutamine (polyQ) tracts are a pathological hallmark of Huntington's disease (HD). The molecular mechanisms by which these structures are formed and cause neuronal dysfunction and toxicity are poorly understood. Here, we utilized available gene expression data sets of selected brain regions of HD patients and controls for systematic interaction network filtering in order to predict disease-relevant, brain region-specific HTT interaction partners. Starting from a large protein-protein interaction (PPI) data set, a step-by-step computational filtering strategy facilitated the generation of a focused PPI network that directly or indirectly connects 13 proteins potentially dysregulated in HD with the disease protein HTT. This network enabled the discovery of the neuron-specific protein CRMP1 that targets aggregation-prone, N-terminal HTT fragments and suppresses their spontaneous self-assembly into proteotoxic structures in various models of HD. Experimental validation indicates that our network filtering procedure provides a simple but powerful strategy to identify disease-relevant proteins that influence misfolding and aggregation of polyQ disease proteins.
© 2015 Stroedicke et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


Similar articles
-
The emerging role of the first 17 amino acids of huntingtin in Huntington's disease.Biomol Concepts. 2015 Mar;6(1):33-46. doi: 10.1515/bmc-2015-0001. Biomol Concepts. 2015. PMID: 25741791 Free PMC article. Review.
-
Spontaneous self-assembly of pathogenic huntingtin exon 1 protein into amyloid structures.Essays Biochem. 2014;56:167-80. doi: 10.1042/bse0560167. Essays Biochem. 2014. PMID: 25131594 Review.
-
Green tea (-)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington's disease models.Hum Mol Genet. 2006 Sep 15;15(18):2743-51. doi: 10.1093/hmg/ddl210. Epub 2006 Aug 7. Hum Mol Genet. 2006. PMID: 16893904
-
14-3-3zeta is indispensable for aggregate formation of polyglutamine-expanded huntingtin protein.Neurosci Lett. 2008 Jan 24;431(1):45-50. doi: 10.1016/j.neulet.2007.11.018. Epub 2007 Nov 17. Neurosci Lett. 2008. PMID: 18078716
-
Cholesterol Modifies Huntingtin Binding to, Disruption of, and Aggregation on Lipid Membranes.Biochemistry. 2016 Jan 12;55(1):92-102. doi: 10.1021/acs.biochem.5b00900. Epub 2015 Dec 22. Biochemistry. 2016. PMID: 26652744 Free PMC article.
Cited by
-
Spatiotemporal Proteomic Profiling of Huntington's Disease Inclusions Reveals Widespread Loss of Protein Function.Cell Rep. 2017 Nov 21;21(8):2291-2303. doi: 10.1016/j.celrep.2017.10.097. Cell Rep. 2017. PMID: 29166617 Free PMC article.
-
Integrative Proteome Analysis Revels 3-Hydroxybutyrate Exerts Neuroprotective Effect by Influencing Chromatin Bivalency.Int J Mol Sci. 2023 Jan 3;24(1):868. doi: 10.3390/ijms24010868. Int J Mol Sci. 2023. PMID: 36614311 Free PMC article.
-
On the identification of potential novel therapeutic targets for spinocerebellar ataxia type 1 (SCA1) neurodegenerative disease using EvoPPI3.J Integr Bioinform. 2023 Feb 28;20(2):20220056. doi: 10.1515/jib-2022-0056. eCollection 2023 Jun 1. J Integr Bioinform. 2023. PMID: 36848492 Free PMC article.
-
Protein misassembly and aggregation as potential convergence points for non-genetic causes of chronic mental illness.Mol Psychiatry. 2019 Jul;24(7):936-951. doi: 10.1038/s41380-018-0133-2. Epub 2018 Aug 8. Mol Psychiatry. 2019. PMID: 30089789 Review.
-
Phosphorylated CRMP1, axon guidance protein, is a component of spheroids and is involved in axonal pathology in amyotrophic lateral sclerosis.Front Neurol. 2022 Sep 27;13:994676. doi: 10.3389/fneur.2022.994676. eCollection 2022. Front Neurol. 2022. PMID: 36237616 Free PMC article.
References
-
- Abraham VC, Taylor DL, Haskins JR. 2004. High content screening applied to large-scale cell biology. Trends Biotechnol 22: 15–22. - PubMed
-
- Apostol BL, Illes K, Pallos J, Bodai L, Wu J, Strand A, Schweitzer ES, Olson JM, Kazantsev A, Marsh JL, et al.2006. Mutant huntingtin alters MAPK signaling pathways in PC12 and striatal cells: ERK1/2 protects against mutant huntingtin-associated toxicity. Hum Mol Genet 15: 273–285. - PubMed
-
- Aylward EH. 2007. Change in MRI striatal volumes as a biomarker in preclinical Huntington's disease. Brain Res Bull 72: 152–158. - PubMed
-
- Behrends C, Langer CA, Boteva R, Böttcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, et al.2006. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell 23: 887–897. - PubMed
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 57: 289–300.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
