Unbiased Cell-based Screening in a Neuronal Cell Model of Batten Disease Highlights an Interaction between Ca2+ Homeostasis, Autophagy, and CLN3 Protein Function
- PMID: 25878248
- PMCID: PMC4505505
- DOI: 10.1074/jbc.M114.621706
Unbiased Cell-based Screening in a Neuronal Cell Model of Batten Disease Highlights an Interaction between Ca2+ Homeostasis, Autophagy, and CLN3 Protein Function
Abstract
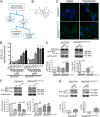
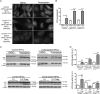
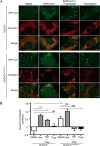
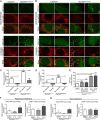
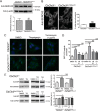
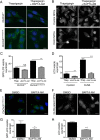
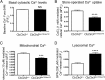
Abnormal accumulation of undigested macromolecules, often disease-specific, is a major feature of lysosomal and neurodegenerative disease and is frequently attributed to defective autophagy. The mechanistic underpinnings of the autophagy defects are the subject of intense research, which is aided by genetic disease models. To gain an improved understanding of the pathways regulating defective autophagy specifically in juvenile neuronal ceroid lipofuscinosis (JNCL or Batten disease), a neurodegenerative disease of childhood, we developed and piloted a GFP-microtubule-associated protein 1 light chain 3 (GFP-LC3) screening assay to identify, in an unbiased fashion, genotype-sensitive small molecule autophagy modifiers, employing a JNCL neuronal cell model bearing the most common disease mutation in CLN3. Thapsigargin, a sarco/endoplasmic reticulum Ca(2+)-ATPase (SERCA) Ca(2+) pump inhibitor, reproducibly displayed significantly more activity in the mouse JNCL cells, an effect that was also observed in human-induced pluripotent stem cell-derived JNCL neural progenitor cells. The mechanism of thapsigargin sensitivity was Ca(2+)-mediated, and autophagosome accumulation in JNCL cells could be reversed by Ca(2+) chelation. Interrogation of intracellular Ca(2+) handling highlighted alterations in endoplasmic reticulum, mitochondrial, and lysosomal Ca(2+) pools and in store-operated Ca(2+) uptake in JNCL cells. These results further support an important role for the CLN3 protein in intracellular Ca(2+) handling and in autophagic pathway flux and establish a powerful new platform for therapeutic screening.
Keywords: CLN3; GFP-LC3; autophagy; calcium; chemical biology; endoplasmic reticulum (ER); juvenile neuronal ceroid lipofuscinosis (JNCL); lysosomal storage disease; lysosome; thapsigargin.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Cln3-mutations underlying juvenile neuronal ceroid lipofuscinosis cause significantly reduced levels of Palmitoyl-protein thioesterases-1 (Ppt1)-protein and Ppt1-enzyme activity in the lysosome.J Inherit Metab Dis. 2019 Sep;42(5):944-954. doi: 10.1002/jimd.12106. Epub 2019 May 14. J Inherit Metab Dis. 2019. PMID: 31025705 Free PMC article.
-
An Autophagy Modifier Screen Identifies Small Molecules Capable of Reducing Autophagosome Accumulation in a Model of CLN3-Mediated Neurodegeneration.Cells. 2019 Nov 27;8(12):1531. doi: 10.3390/cells8121531. Cells. 2019. PMID: 31783699 Free PMC article.
-
Loss of CLN3, the gene mutated in juvenile neuronal ceroid lipofuscinosis, leads to metabolic impairment and autophagy induction in retinal pigment epithelium.Biochim Biophys Acta Mol Basis Dis. 2020 Oct 1;1866(10):165883. doi: 10.1016/j.bbadis.2020.165883. Epub 2020 Jun 25. Biochim Biophys Acta Mol Basis Dis. 2020. PMID: 32592935 Free PMC article.
-
[Mechanisms of juvenile neuronal ceroid lipofuscinosis (JNCL)].Yi Chuan. 2009 Aug;31(8):779-84. doi: 10.3724/sp.j.1005.2009.00779. Yi Chuan. 2009. PMID: 19689937 Review. Chinese.
-
The CLN3 gene and protein: What we know.Mol Genet Genomic Med. 2019 Dec;7(12):e859. doi: 10.1002/mgg3.859. Epub 2019 Sep 30. Mol Genet Genomic Med. 2019. PMID: 31568712 Free PMC article. Review.
Cited by
-
Astrocytes and lysosomal storage diseases.Neuroscience. 2016 May 26;323:195-206. doi: 10.1016/j.neuroscience.2015.05.061. Epub 2015 May 30. Neuroscience. 2016. PMID: 26037807 Free PMC article. Review.
-
Asthma Susceptibility Gene ORMDL3 Promotes Autophagy in Human Bronchial Epithelium.Am J Respir Cell Mol Biol. 2022 Jun;66(6):661-670. doi: 10.1165/rcmb.2021-0305OC. Am J Respir Cell Mol Biol. 2022. PMID: 35353673 Free PMC article.
-
Converging roles of PSENEN/PEN2 and CLN3 in the autophagy-lysosome system.Autophagy. 2022 Sep;18(9):2068-2085. doi: 10.1080/15548627.2021.2016232. Epub 2021 Dec 29. Autophagy. 2022. PMID: 34964690 Free PMC article.
-
An iPSC-Derived Neuron Model of CLN3 Disease Facilitates Small Molecule Phenotypic Screening.ACS Pharmacol Transl Sci. 2020 Sep 1;3(5):931-947. doi: 10.1021/acsptsci.0c00077. eCollection 2020 Oct 9. ACS Pharmacol Transl Sci. 2020. PMID: 33073192 Free PMC article.
-
Lysosomal Ca2+ Homeostasis and Signaling in Health and Disease.Cold Spring Harb Perspect Biol. 2020 Jun 1;12(6):a035311. doi: 10.1101/cshperspect.a035311. Cold Spring Harb Perspect Biol. 2020. PMID: 31653642 Free PMC article. Review.
References
-
- Mole S. E., Williams R. E., Goebel H. H. (eds) (2011) The Neuronal Ceroid Lipofuscinoses (Batten Disease), 2nd Ed., Oxford University Press, Oxford, UK
-
- Kollmann K., Uusi-Rauva K., Scifo E., Tyynelä J., Jalanko A., Braulke T. (2013) Cell biology and function of neuronal ceroid lipofuscinosis-related proteins. Biochim. Biophys. Acta 1832, 1866–1881 - PubMed
-
- Warrier V., Vieira M., Mole S. E. (2013) Genetic basis and phenotypic correlations of the neuronal ceroid lipofusinoses. Biochim. Biophys. Acta 1832, 1827–1830 - PubMed
-
- International Batten Disease Consortium (1995) Isolation of a novel gene underlying Batten disease, CLN3. The International Batten Disease Consortium. Cell 82, 949–957 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

