Human Cytomegalovirus UL135 and UL136 Genes Are Required for Postentry Tropism in Endothelial Cells
- PMID: 25878111
- PMCID: PMC4468499
- DOI: 10.1128/JVI.00284-15
Human Cytomegalovirus UL135 and UL136 Genes Are Required for Postentry Tropism in Endothelial Cells
Abstract
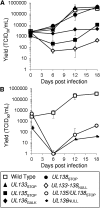
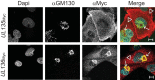
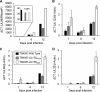
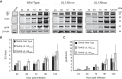
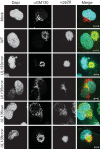
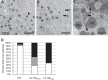
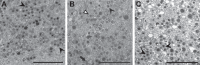
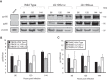
Endothelial cells (ECs) are a critical target of viruses, and infection of the endothelium represents a defining point in viral pathogenesis. Human cytomegalovirus (HCMV), the prototypical betaherpesvirus, encodes proteins specialized for entry into ECs and delivery of the genome to the nuclei of ECs. Virus strains competent to enter ECs replicate with differing efficiencies, suggesting that the virus encodes genes for postentry tropism in ECs. We previously reported a specific requirement for the UL133/8 locus of HCMV for replication in ECs. The UL133/8 locus harbors four genes: UL133, UL135, UL136, and UL138. In this study, we find that while UL133 and UL138 are dispensable for replication in ECs, both UL135 and UL136 are important. These genes are not required for virus entry or the expression of viral genes. The phenotypes associated with disruption of either gene reflect phenotypes observed for the UL133/8NULL virus, which lacks the entire UL133/8 locus, but are largely distinct from one another. Viruses lacking UL135 fail to properly envelop capsids in the cytoplasm, produce fewer dense bodies (DB) than the wild-type (WT) virus, and are unable to incorporate viral products into multivesicular bodies (MVB). Viruses lacking UL136 also fail to properly envelop virions and produce larger dense bodies than the WT virus. Our results indicate roles for the UL135 and UL136 proteins in commandeering host membrane-trafficking pathways for virus maturation. UL135 and UL136 represent the first HCMV genes crucial for early- to late-stage tropism in ECs.
Importance: Human cytomegalovirus (HCMV) persists in the majority of the world's population. While typically asymptomatic in healthy hosts, HCMV can cause significant morbidity and mortality in immunocompromised or naïve individuals, particularly transplant patients and patients with congenital infections, respectively. Lifelong persistence of the virus may also contribute to age-related pathologies, such as vascular disease. One aspect of HCMV infection contributing to complex and varied pathogenesis is the diverse array of cell types that this virus infects in the host. The vascular endothelium is a particularly important target of infection, contributing to viral dissemination and likely leading to CMV complications following transplantation. In this work, we identify two viral gene products required for postentry tropism in endothelial cells. Identifying tropism factors required for replication in critical cell targets of infection is important for the development of strategies to restrict virus replication.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
The Role of the Human Cytomegalovirus UL133-UL138 Gene Locus in Latency and Reactivation.Viruses. 2020 Jul 1;12(7):714. doi: 10.3390/v12070714. Viruses. 2020. PMID: 32630219 Free PMC article. Review.
-
Complex Interplay of the UL136 Isoforms Balances Cytomegalovirus Replication and Latency.mBio. 2016 Mar 1;7(2):e01986. doi: 10.1128/mBio.01986-15. mBio. 2016. PMID: 26933055 Free PMC article.
-
An endothelial cell-specific requirement for the UL133-UL138 locus of human cytomegalovirus for efficient virus maturation.J Virol. 2013 Mar;87(6):3062-75. doi: 10.1128/JVI.02510-12. Epub 2013 Jan 2. J Virol. 2013. PMID: 23283945 Free PMC article.
-
Antagonistic determinants controlling replicative and latent states of human cytomegalovirus infection.J Virol. 2014 Jun;88(11):5987-6002. doi: 10.1128/JVI.03506-13. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623432 Free PMC article.
-
The power of human cytomegalovirus (HCMV) hijacked UL/b' functions lost in vitro.Acta Virol. 2020;64(2):117-130. doi: 10.4149/av_2020_202. Acta Virol. 2020. PMID: 32551781 Review.
Cited by
-
The Role of the Human Cytomegalovirus UL133-UL138 Gene Locus in Latency and Reactivation.Viruses. 2020 Jul 1;12(7):714. doi: 10.3390/v12070714. Viruses. 2020. PMID: 32630219 Free PMC article. Review.
-
Complex Interplay of the UL136 Isoforms Balances Cytomegalovirus Replication and Latency.mBio. 2016 Mar 1;7(2):e01986. doi: 10.1128/mBio.01986-15. mBio. 2016. PMID: 26933055 Free PMC article.
-
Cell type-specific biogenesis of novel vesicles containing viral products in human cytomegalovirus infection.J Virol. 2021 May 10;95(11):e02358-20. doi: 10.1128/JVI.02358-20. Epub 2021 Mar 24. J Virol. 2021. PMID: 33762413 Free PMC article.
-
Human Cytomegalovirus UL135 Interacts with Host Adaptor Proteins To Regulate Epidermal Growth Factor Receptor and Reactivation from Latency.J Virol. 2018 Sep 26;92(20):e00919-18. doi: 10.1128/JVI.00919-18. Print 2018 Oct 15. J Virol. 2018. PMID: 30089695 Free PMC article.
-
Cytomegalovirus Latency and Reactivation: An Intricate Interplay With the Host Immune Response.Front Cell Infect Microbiol. 2020 Mar 31;10:130. doi: 10.3389/fcimb.2020.00130. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32296651 Free PMC article. Review.
References
-
- Jarvis MA, Nelson JA. 2007. Molecular basis of persistence and latency, chapter 42 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. - PubMed
-
- Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2701–2673. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

