Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming
- PMID: 25877919
- PMCID: PMC4403257
- DOI: 10.1101/gad.255109.114
Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming
Erratum in
-
Corrigendum: Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming.Genes Dev. 2015 Jun 15;29(12):1341. doi: 10.1101/gad.266601.115. Genes Dev. 2015. PMID: 26109054 Free PMC article. No abstract available.
Abstract
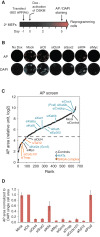
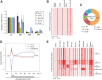
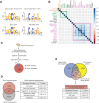
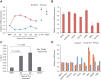
Embryonic stem cells are maintained in a self-renewing and pluripotent state by multiple regulatory pathways. Pluripotent-specific transcriptional networks are sequentially reactivated as somatic cells reprogram to achieve pluripotency. How epigenetic regulators modulate this process and contribute to somatic cell reprogramming is not clear. Here we performed a functional RNAi screen to identify the earliest epigenetic regulators required for reprogramming. We identified components of the SAGA histone acetyltransferase complex, in particular Gcn5, as critical regulators of reprogramming initiation. Furthermore, we showed in mouse pluripotent stem cells that Gcn5 strongly associates with Myc and that, upon initiation of somatic reprogramming, Gcn5 and Myc form a positive feed-forward loop that activates a distinct alternative splicing network and the early acquisition of pluripotency-associated splicing events. These studies expose a Myc-SAGA pathway that drives expression of an essential alternative splicing regulatory network during somatic cell reprogramming.
Keywords: Gcn5; Myc; SAGA; alternative splicing; iPSCs; reprogramming.
© 2015 Hirsch et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
Develop-WNTs in somatic cell reprogramming.Cell Stem Cell. 2008 Nov 6;3(5):465-6. doi: 10.1016/j.stem.2008.10.011. Cell Stem Cell. 2008. PMID: 18983957
-
Multifaceted regulation of somatic cell reprogramming by mRNA translational control.Cell Stem Cell. 2014 May 1;14(5):606-16. doi: 10.1016/j.stem.2014.02.005. Epub 2014 Mar 13. Cell Stem Cell. 2014. PMID: 24630793
-
Transcriptome Signature and Regulation in Human Somatic Cell Reprogramming.Stem Cell Reports. 2015 Jun 9;4(6):1125-39. doi: 10.1016/j.stemcr.2015.04.009. Epub 2015 May 21. Stem Cell Reports. 2015. PMID: 26004630 Free PMC article.
-
Ground rules of the pluripotency gene regulatory network.Nat Rev Genet. 2017 Mar;18(3):180-191. doi: 10.1038/nrg.2016.156. Epub 2017 Jan 3. Nat Rev Genet. 2017. PMID: 28045100 Review.
-
Mechanism of Induction: Induced Pluripotent Stem Cells (iPSCs).J Stem Cells. 2015;10(1):43-62. J Stem Cells. 2015. PMID: 26665937 Review.
Cited by
-
Dynamic modules of the coactivator SAGA in eukaryotic transcription.Exp Mol Med. 2020 Jul;52(7):991-1003. doi: 10.1038/s12276-020-0463-4. Epub 2020 Jul 3. Exp Mol Med. 2020. PMID: 32616828 Free PMC article. Review.
-
GCN5 Regulates FGF Signaling and Activates Selective MYC Target Genes during Early Embryoid Body Differentiation.Stem Cell Reports. 2018 Jan 9;10(1):287-299. doi: 10.1016/j.stemcr.2017.11.009. Epub 2017 Dec 14. Stem Cell Reports. 2018. PMID: 29249668 Free PMC article.
-
Epigenetic Control of Reprogramming and Transdifferentiation by Histone Modifications.Stem Cell Rev Rep. 2016 Dec;12(6):708-720. doi: 10.1007/s12015-016-9682-4. Stem Cell Rev Rep. 2016. PMID: 27623868 Review.
-
Single cell RNA-seq identifies the origins of heterogeneity in efficient cell transdifferentiation and reprogramming.Elife. 2019 Mar 12;8:e41627. doi: 10.7554/eLife.41627. Elife. 2019. PMID: 30860479 Free PMC article.
-
TAF5L and TAF6L Maintain Self-Renewal of Embryonic Stem Cells via the MYC Regulatory Network.Mol Cell. 2019 Jun 20;74(6):1148-1163.e7. doi: 10.1016/j.molcel.2019.03.025. Epub 2019 Apr 17. Mol Cell. 2019. PMID: 31005419 Free PMC article.
References
-
- Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, Andrews PW. 2008. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells 26: 3068–3074. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
