Personalized genomic analyses for cancer mutation discovery and interpretation
- PMID: 25877891
- PMCID: PMC4442685
- DOI: 10.1126/scitranslmed.aaa7161
Personalized genomic analyses for cancer mutation discovery and interpretation
Abstract
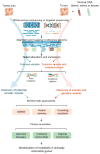
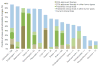
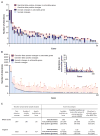
Massively parallel sequencing approaches are beginning to be used clinically to characterize individual patient tumors and to select therapies based on the identified mutations. A major question in these analyses is the extent to which these methods identify clinically actionable alterations and whether the examination of the tumor tissue alone is sufficient or whether matched normal DNA should also be analyzed to accurately identify tumor-specific (somatic) alterations. To address these issues, we comprehensively evaluated 815 tumor-normal paired samples from patients of 15 tumor types. We identified genomic alterations using next-generation sequencing of whole exomes or 111 targeted genes that were validated with sensitivities >95% and >99%, respectively, and specificities >99.99%. These analyses revealed an average of 140 and 4.3 somatic mutations per exome and targeted analysis, respectively. More than 75% of cases had somatic alterations in genes associated with known therapies or current clinical trials. Analyses of matched normal DNA identified germline alterations in cancer-predisposing genes in 3% of patients with apparently sporadic cancers. In contrast, a tumor-only sequencing approach could not definitively identify germline changes in cancer-predisposing genes and led to additional false-positive findings comprising 31% and 65% of alterations identified in targeted and exome analyses, respectively, including in potentially actionable genes. These data suggest that matched tumor-normal sequencing analyses are essential for precise identification and interpretation of somatic and germline alterations and have important implications for the diagnostic and therapeutic management of cancer patients.
Copyright © 2015, American Association for the Advancement of Science.
Conflict of interest statement
Figures

Comment in
-
Genomic sequencing of only tumor tissue could be misleading in nearly half of patients, study shows.BMJ. 2015 Apr 16;350:h2036. doi: 10.1136/bmj.h2036. BMJ. 2015. PMID: 25882003 No abstract available.
Similar articles
-
A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal.PLoS Comput Biol. 2018 Feb 7;14(2):e1005965. doi: 10.1371/journal.pcbi.1005965. eCollection 2018 Feb. PLoS Comput Biol. 2018. PMID: 29415044 Free PMC article.
-
Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response.JAMA Oncol. 2015 Jul;1(4):466-74. doi: 10.1001/jamaoncol.2015.1313. JAMA Oncol. 2015. PMID: 26181256 Free PMC article.
-
The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine.Genome Med. 2016 Jul 26;8(1):79. doi: 10.1186/s13073-016-0333-9. Genome Med. 2016. PMID: 27460824 Free PMC article.
-
Whole-exome sequencing reveals recurrent somatic mutation networks in cancer.Cancer Lett. 2013 Nov 1;340(2):270-6. doi: 10.1016/j.canlet.2012.11.002. Epub 2012 Nov 12. Cancer Lett. 2013. PMID: 23153794 Review.
-
The emerging significance of secondary germline testing in cancer genomics.J Pathol. 2018 Apr;244(5):610-615. doi: 10.1002/path.5031. Epub 2018 Feb 16. J Pathol. 2018. PMID: 29293272 Review.
Cited by
-
TumorNext: A comprehensive tumor profiling assay that incorporates high resolution copy number analysis and germline status to improve testing accuracy.Oncotarget. 2016 Oct 18;7(42):68206-68228. doi: 10.18632/oncotarget.11910. Oncotarget. 2016. PMID: 27626691 Free PMC article.
-
Next-generation sequencing to guide cancer therapy.Genome Med. 2015 Jul 29;7(1):80. doi: 10.1186/s13073-015-0203-x. eCollection 2015. Genome Med. 2015. PMID: 26221189 Free PMC article. Review.
-
Genomic Signatures from Clinical Tumor Sequencing in Patients with Breast Cancer Having Germline BRCA1/2 Mutation.Cancer Res Treat. 2023 Jan;55(1):155-166. doi: 10.4143/crt.2021.1567. Epub 2022 Jun 8. Cancer Res Treat. 2023. PMID: 35681111 Free PMC article.
-
Differential metabolic activity and discovery of therapeutic targets using summarized metabolic pathway models.NPJ Syst Biol Appl. 2019 Mar 1;5:7. doi: 10.1038/s41540-019-0087-2. eCollection 2019. NPJ Syst Biol Appl. 2019. PMID: 30854222 Free PMC article.
-
Reliable Analysis of Clinical Tumor-Only Whole-Exome Sequencing Data.JCO Clin Cancer Inform. 2020 Apr;4:321-335. doi: 10.1200/CCI.19.00130. JCO Clin Cancer Inform. 2020. PMID: 32282230 Free PMC article.
References
-
- Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, Jane-Valbuena J, Friedrich DC, Kryukov G, Carter SL, McKenna A, Sivachenko A, Rosenberg M, Kiezun A, Voet D, Lawrence M, Lichtenstein LT, Gentry JG, Huang FW, Fostel J, Farlow D, Barbie D, Gandhi L, Lander ES, Gray SW, Joffe S, Janne P, Garber J, MacConaill L, Lindeman N, Rollins B, Kantoff P, Fisher SA, Gabriel S, Getz G, Garraway LA. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–688. - PMC - PubMed
-
- Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, Sun J, Juhn F, Brennan K, Iwanik K, Maillet A, Buell J, White E, Zhao M, Balasubramanian S, Terzic S, Richards T, Banning V, Garcia L, Mahoney K, Zwirko Z, Donahue A, Beltran H, Mosquera JM, Rubin MA, Dogan S, Hedvat CV, Berger MF, Pusztai L, Lechner M, Boshoff C, Jarosz M, Vietz C, Parker A, Miller VA, Ross JS, Curran J, Cronin MT, Stephens PJ, Lipson D, Yelensky R. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. - PMC - PubMed
-
- Garraway LA, Jänne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2012;2:214–226. - PubMed
-
- Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, Ducar M, Van Hummelen P, Macconaill LE, Hahn WC, Meyerson M, Gabriel SB, Garraway LA. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. - PMC - PubMed
-
- Dias-Santagata D, Akhavanfard S, David SS, Vernovsky K, Kuhlmann G, Boisvert SL, Stubbs H, McDermott U, Settleman J, Kwak EL, Clark JW, Isakoff SJ, Sequist LV, Engelman JA, Lynch TJ, Haber DA, Louis DN, Ellisen LW, Borger DR, Iafrate AJ. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

