Human cytomegalovirus pUL97 kinase induces global changes in the infected cell phosphoproteome
- PMID: 25867546
- PMCID: PMC4522931
- DOI: 10.1002/pmic.201400607
Human cytomegalovirus pUL97 kinase induces global changes in the infected cell phosphoproteome
Abstract
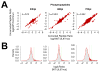
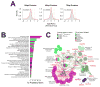
Replication of human cytomegalovirus (HCMV) is regulated in part by cellular kinases and the single viral Ser/Thr kinase, pUL97. The virus-coded kinase augments the replication of HCMV by enabling nuclear egress and altering cell cycle progression. These roles are accomplished through direct phosphorylation of nuclear lamins and the retinoblastoma protein, respectively. In an effort to identify additional pUL97 substrates, we analyzed the phosphoproteome of SILAC-labeled human fibroblasts during infection with either wild-type HCMV or a pUL97 kinase-dead mutant virus. Phosphopeptides were enriched over a titanium dioxide matrix and analyzed by high-resolution MS. We identified 157 unambiguous phosphosites from 106 cellular and 17 viral proteins whose phosphorylation required UL97. Analysis of peptides containing these sites allowed the identification of several candidate pUL97 phosphorylation motifs, including a completely novel phosphorylation motif, LxSP. Substrates harboring the LxSP motif were enriched in nucleocytoplasmic transport functions, including a number of components of the nuclear pore complex. These results extend the known functions of pUL97 and suggest that modulation of nuclear pore function may be important during HCMV replication.
Keywords: Cell biology; Herpesvirus; Human cytomegalovirus; Nuclear pore complex; TREX complex; Viral kinase.
© 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Regulatory roles of protein kinases in cytomegalovirus replication.Adv Virus Res. 2011;80:69-101. doi: 10.1016/B978-0-12-385987-7.00004-X. Adv Virus Res. 2011. PMID: 21762822 Review.
-
Proteomic identification of nuclear processes manipulated by cytomegalovirus early during infection.Proteomics. 2015 Jun;15(12):1995-2005. doi: 10.1002/pmic.201400599. Epub 2015 Apr 28. Proteomics. 2015. PMID: 25758553 Free PMC article.
-
Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus.J Biol Chem. 2010 Apr 30;285(18):13979-89. doi: 10.1074/jbc.M109.063628. Epub 2010 Mar 4. J Biol Chem. 2010. PMID: 20202933 Free PMC article.
-
Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina.J Biol Chem. 2005 Sep 30;280(39):33357-67. doi: 10.1074/jbc.M502672200. Epub 2005 Jun 23. J Biol Chem. 2005. PMID: 15975922
-
Understanding the Cytomegalovirus Cyclin-Dependent Kinase Ortholog pUL97 as a Multifaceted Regulator and an Antiviral Drug Target.Cells. 2024 Aug 13;13(16):1338. doi: 10.3390/cells13161338. Cells. 2024. PMID: 39195228 Free PMC article. Review.
Cited by
-
Advances in the Development of Therapeutics for Cytomegalovirus Infections.J Infect Dis. 2020 Mar 5;221(Suppl 1):S32-S44. doi: 10.1093/infdis/jiz493. J Infect Dis. 2020. PMID: 32134483 Free PMC article. Review.
-
Conserved Herpesvirus Protein Kinases Target SAMHD1 to Facilitate Virus Replication.Cell Rep. 2019 Jul 9;28(2):449-459.e5. doi: 10.1016/j.celrep.2019.04.020. Cell Rep. 2019. PMID: 31291580 Free PMC article.
-
Phosphoproteomic Profiling Reveals Epstein-Barr Virus Protein Kinase Integration of DNA Damage Response and Mitotic Signaling.PLoS Pathog. 2015 Dec 29;11(12):e1005346. doi: 10.1371/journal.ppat.1005346. eCollection 2015 Dec. PLoS Pathog. 2015. PMID: 26714015 Free PMC article.
-
Functional Relevance of the Interaction between Human Cyclins and the Cytomegalovirus-Encoded CDK-Like Protein Kinase pUL97.Viruses. 2021 Jun 27;13(7):1248. doi: 10.3390/v13071248. Viruses. 2021. PMID: 34198986 Free PMC article.
-
Proteomic Interaction Patterns between Human Cyclins, the Cyclin-Dependent Kinase Ortholog pUL97 and Additional Cytomegalovirus Proteins.Viruses. 2016 Aug 18;8(8):219. doi: 10.3390/v8080219. Viruses. 2016. PMID: 27548200 Free PMC article.
References
-
- Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams and Wilkins; Philadelphia: 2007. pp. 2701–2772.
-
- Muganda-Ojiaku PM, Huang ES. Alteration of protein phosphorylation patterns in cell lines morphologically transformed by human cytomegalovirus. Cancer Biochem Biophys. 1987;9:179–189. - PubMed
-
- Muranyi W, Haas J, Wagner M, Krohne G, Koszinowski UH. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science. 2002;297:854–857. - PubMed
-
- Sun B, Harrowe G, Reinhard C, Yoshihara C, et al. Modulation of human cytomegalovirus immediate-early gene enhancer by mitogen-activated protein kinase kinase kinase-1. J Cell Biochem. 2001;83:563–573. - PubMed
-
- Milbradt J, Auerochs S, Marschall M. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J Gen Virol. 2007;88:2642–2650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

