Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets
- PMID: 25855536
- PMCID: PMC4403382
- DOI: 10.1038/ncomms7744
Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets
Abstract
Pancreatic ductal adenocarcinoma (PDA) has a dismal prognosis and insights into both disease etiology and targeted intervention are needed. A total of 109 micro-dissected PDA cases were subjected to whole-exome sequencing. Microdissection enriches tumour cellularity and enhances mutation calling. Here we show that environmental stress and alterations in DNA repair genes associate with distinct mutation spectra. Copy number alterations target multiple tumour suppressive/oncogenic loci; however, amplification of MYC is uniquely associated with poor outcome and adenosquamous subtype. We identify multiple novel mutated genes in PDA, with select genes harbouring prognostic significance. RBM10 mutations associate with longer survival in spite of histological features of aggressive disease. KRAS mutations are observed in >90% of cases, but codon Q61 alleles are selectively associated with improved survival. Oncogenic BRAF mutations are mutually exclusive with KRAS and define sensitivity to vemurafenib in PDA models. High-frequency alterations in Wnt signalling, chromatin remodelling, Hedgehog signalling, DNA repair and cell cycle processes are observed. Together, these data delineate new genetic diversity of PDA and provide insights into prognostic determinants and therapeutic targets.
Figures

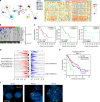
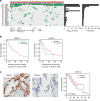


Comment in
-
Pancreatic cancer: Targeting KRAS and the vitamin D receptor via microtubules.Nat Rev Clin Oncol. 2015 Aug;12(8):442-4. doi: 10.1038/nrclinonc.2015.125. Epub 2015 Jul 14. Nat Rev Clin Oncol. 2015. PMID: 26169922 No abstract available.
Similar articles
-
Clinical study of genomic drivers in pancreatic ductal adenocarcinoma.Br J Cancer. 2017 Aug 8;117(4):572-582. doi: 10.1038/bjc.2017.209. Epub 2017 Jul 18. Br J Cancer. 2017. PMID: 28720843 Free PMC article.
-
Genomic and Epigenomic Landscaping Defines New Therapeutic Targets for Adenosquamous Carcinoma of the Pancreas.Cancer Res. 2020 Oct 15;80(20):4324-4334. doi: 10.1158/0008-5472.CAN-20-0078. Epub 2020 Sep 14. Cancer Res. 2020. PMID: 32928922 Free PMC article.
-
KRAS mutations in codon 12 or 13 are associated with worse prognosis in pancreatic ductal adenocarcinoma.Pancreas. 2014 May;43(4):578-83. doi: 10.1097/MPA.0000000000000077. Pancreas. 2014. PMID: 24681874
-
Clinicopathological Implications of Wingless/int1 (WNT) Signaling Pathway in Pancreatic Ductal Adenocarcinoma.J UOEH. 2016 Mar 1;38(1):1-8. doi: 10.7888/juoeh.38.1. J UOEH. 2016. PMID: 26972939 Review.
-
KRAS-related proteins in pancreatic cancer.Pharmacol Ther. 2016 Dec;168:29-42. doi: 10.1016/j.pharmthera.2016.09.003. Epub 2016 Sep 3. Pharmacol Ther. 2016. PMID: 27595930 Review.
Cited by
-
RBM10 Regulates Tumor Apoptosis, Proliferation, and Metastasis.Front Oncol. 2021 Feb 24;11:603932. doi: 10.3389/fonc.2021.603932. eCollection 2021. Front Oncol. 2021. PMID: 33718153 Free PMC article. Review.
-
TLR2 and TLR9 Blockade Using Specific Intrabodies Inhibits Inflammation-Mediated Pancreatic Cancer Cell Growth.Antibodies (Basel). 2024 Feb 1;13(1):11. doi: 10.3390/antib13010011. Antibodies (Basel). 2024. PMID: 38390872 Free PMC article.
-
Complete response to talazoparib in patient with pancreatic adenocarcinoma harboring somatic PALB2 mutation: A case report and literature review.Front Oncol. 2022 Sep 2;12:953908. doi: 10.3389/fonc.2022.953908. eCollection 2022. Front Oncol. 2022. PMID: 36119518 Free PMC article.
-
Hedgehog pathway overexpression in pancreatic cancer is abrogated by new-generation taxoid SB-T-1216.Pharmacogenomics J. 2017 Oct;17(5):452-460. doi: 10.1038/tpj.2016.55. Epub 2016 Aug 30. Pharmacogenomics J. 2017. PMID: 27573236
-
Identification of CD318, TSPAN8 and CD66c as target candidates for CAR T cell based immunotherapy of pancreatic adenocarcinoma.Nat Commun. 2021 Mar 5;12(1):1453. doi: 10.1038/s41467-021-21774-4. Nat Commun. 2021. PMID: 33674603 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

