Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal
- PMID: 25808495
- PMCID: PMC4436827
- DOI: 10.1091/mbc.E14-11-1500
Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal
Abstract
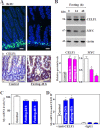
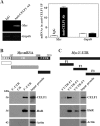
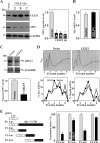
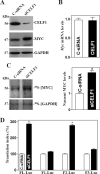
The mammalian intestinal epithelium is one of the most rapidly self-renewing tissues in the body, and its integrity is preserved through strict regulation. The RNA-binding protein (RBP) ELAV-like family member 1 (CELF1), also referred to as CUG-binding protein 1 (CUGBP1), regulates the stability and translation of target mRNAs and is implicated in many aspects of cellular physiology. We show that CELF1 competes with the RBP HuR to modulate MYC translation and regulates intestinal epithelial homeostasis. Growth inhibition of the small intestinal mucosa by fasting in mice was associated with increased CELF1/Myc mRNA association and decreased MYC expression. At the molecular level, CELF1 was found to bind the 3'-untranslated region (UTR) of Myc mRNA and repressed MYC translation without affecting total Myc mRNA levels. HuR interacted with the same Myc 3'-UTR element, and increasing the levels of HuR decreased CELF1 binding to Myc mRNA. In contrast, increasing the concentrations of CELF1 inhibited formation of the [HuR/Myc mRNA] complex. Depletion of cellular polyamines also increased CELF1 and enhanced CELF1 association with Myc mRNA, thus suppressing MYC translation. Moreover, ectopic CELF1 overexpression caused G1-phase growth arrest, whereas CELF1 silencing promoted cell proliferation. These results indicate that CELF1 represses MYC translation by decreasing Myc mRNA association with HuR and provide new insight into the molecular functions of RBPs in the regulation of intestinal mucosal growth.
© 2015 Liu, Ouyang, et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures
Similar articles
-
HuR Enhances Early Restitution of the Intestinal Epithelium by Increasing Cdc42 Translation.Mol Cell Biol. 2017 Mar 17;37(7):e00574-16. doi: 10.1128/MCB.00574-16. Print 2017 Apr 1. Mol Cell Biol. 2017. PMID: 28031329 Free PMC article.
-
CUGBP1 and HuR regulate E-cadherin translation by altering recruitment of E-cadherin mRNA to processing bodies and modulate epithelial barrier function.Am J Physiol Cell Physiol. 2016 Jan 1;310(1):C54-65. doi: 10.1152/ajpcell.00112.2015. Epub 2015 Oct 21. Am J Physiol Cell Physiol. 2016. PMID: 26491048
-
Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation.Mol Biol Cell. 2009 Dec;20(23):4885-98. doi: 10.1091/mbc.e09-07-0550. Epub 2009 Oct 7. Mol Biol Cell. 2009. PMID: 19812253 Free PMC article.
-
Properties of the Regulatory RNA-Binding Protein HuR and its Role in Controlling miRNA Repression.Adv Exp Med Biol. 2011;700:106-23. doi: 10.1007/978-1-4419-7823-3_10. Adv Exp Med Biol. 2011. PMID: 21755477 Review.
-
Properties of the regulatory RNA-binding protein HuR and its role in controlling miRNA repression.Adv Exp Med Biol. 2010;700:106-23. Adv Exp Med Biol. 2010. PMID: 21627034 Review.
Cited by
-
RNA-binding proteins and long noncoding RNAs in intestinal epithelial autophagy and barrier function.Tissue Barriers. 2021 Apr 3;9(2):1895648. doi: 10.1080/21688370.2021.1895648. Epub 2021 Mar 12. Tissue Barriers. 2021. PMID: 33709880 Free PMC article. Review.
-
Correction of RNA-Binding Protein CUGBP1 and GSK3β Signaling as Therapeutic Approach for Congenital and Adult Myotonic Dystrophy Type 1.Int J Mol Sci. 2019 Dec 21;21(1):94. doi: 10.3390/ijms21010094. Int J Mol Sci. 2019. PMID: 31877772 Free PMC article. Review.
-
HuR Enhances Early Restitution of the Intestinal Epithelium by Increasing Cdc42 Translation.Mol Cell Biol. 2017 Mar 17;37(7):e00574-16. doi: 10.1128/MCB.00574-16. Print 2017 Apr 1. Mol Cell Biol. 2017. PMID: 28031329 Free PMC article.
-
AGO2 Mediates MYC mRNA Stability in Hepatocellular Carcinoma.Mol Cancer Res. 2020 Apr;18(4):612-622. doi: 10.1158/1541-7786.MCR-19-0805. Epub 2020 Jan 15. Mol Cancer Res. 2020. PMID: 31941754 Free PMC article.
-
CELF Family Proteins in Cancer: Highlights on the RNA-Binding Protein/Noncoding RNA Regulatory Axis.Int J Mol Sci. 2021 Oct 14;22(20):11056. doi: 10.3390/ijms222011056. Int J Mol Sci. 2021. PMID: 34681716 Free PMC article. Review.
References
-
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. - PubMed
-
- Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6:373–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

