Subsistence strategies in traditional societies distinguish gut microbiomes
- PMID: 25807110
- PMCID: PMC4386023
- DOI: 10.1038/ncomms7505
Subsistence strategies in traditional societies distinguish gut microbiomes
Abstract
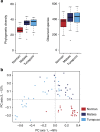
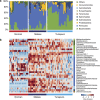
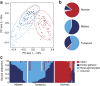
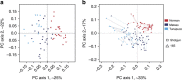
Recent studies suggest that gut microbiomes of urban-industrialized societies are different from those of traditional peoples. Here we examine the relationship between lifeways and gut microbiota through taxonomic and functional potential characterization of faecal samples from hunter-gatherer and traditional agriculturalist communities in Peru and an urban-industrialized community from the US. We find that in addition to taxonomic and metabolic differences between urban and traditional lifestyles, hunter-gatherers form a distinct sub-group among traditional peoples. As observed in previous studies, we find that Treponema are characteristic of traditional gut microbiomes. Moreover, through genome reconstruction (2.2-2.5 MB, coverage depth × 26-513) and functional potential characterization, we discover these Treponema are diverse, fall outside of pathogenic clades and are similar to Treponema succinifaciens, a known carbohydrate metabolizer in swine. Gut Treponema are found in non-human primates and all traditional peoples studied to date, suggesting they are symbionts lost in urban-industrialized societies.
Conflict of interest statement
There are no competing financial interests.
Figures


Similar articles
-
Gut Microbiome of Coexisting BaAka Pygmies and Bantu Reflects Gradients of Traditional Subsistence Patterns.Cell Rep. 2016 Mar 8;14(9):2142-2153. doi: 10.1016/j.celrep.2016.02.013. Epub 2016 Feb 25. Cell Rep. 2016. PMID: 26923597
-
Population structure of human gut bacteria in a diverse cohort from rural Tanzania and Botswana.Genome Biol. 2019 Jan 22;20(1):16. doi: 10.1186/s13059-018-1616-9. Genome Biol. 2019. PMID: 30665461 Free PMC article.
-
A snapshot of gut microbiota of an adult urban population from Western region of India.PLoS One. 2018 Apr 6;13(4):e0195643. doi: 10.1371/journal.pone.0195643. eCollection 2018. PLoS One. 2018. PMID: 29624599 Free PMC article.
-
Modulation of faecal metagenome in Crohn's disease: Role of microRNAs as biomarkers.World J Gastroenterol. 2018 Dec 14;24(46):5223-5233. doi: 10.3748/wjg.v24.i46.5223. World J Gastroenterol. 2018. PMID: 30581271 Free PMC article.
-
Ultra-deep sequencing of Hadza hunter-gatherers recovers vanishing gut microbes.Cell. 2023 Jul 6;186(14):3111-3124.e13. doi: 10.1016/j.cell.2023.05.046. Epub 2023 Jun 21. Cell. 2023. PMID: 37348505 Free PMC article.
Cited by
-
The role of the microbiome in gastrointestinal inflammation.Biosci Rep. 2021 Jun 25;41(6):BSR20203850. doi: 10.1042/BSR20203850. Biosci Rep. 2021. PMID: 34076695 Free PMC article.
-
Potential zoonotic pathogens hosted by endangered bonobos.Sci Rep. 2021 Mar 18;11(1):6331. doi: 10.1038/s41598-021-85849-4. Sci Rep. 2021. PMID: 33737691 Free PMC article.
-
Dysbiotic drift: mental health, environmental grey space, and microbiota.J Physiol Anthropol. 2015 May 7;34(1):23. doi: 10.1186/s40101-015-0061-7. J Physiol Anthropol. 2015. PMID: 25947328 Free PMC article. Review.
-
A 15-day pilot biodiversity intervention with horses in a farm system leads to gut microbiome rewilding in 10 urban Italian children.One Health. 2024 Sep 24;19:100902. doi: 10.1016/j.onehlt.2024.100902. eCollection 2024 Dec. One Health. 2024. PMID: 39399231 Free PMC article.
-
Maternal fecal microbiome predicts gestational age, birth weight and neonatal growth in rural Zimbabwe.EBioMedicine. 2021 Jun;68:103421. doi: 10.1016/j.ebiom.2021.103421. Epub 2021 Jun 15. EBioMedicine. 2021. PMID: 34139432 Free PMC article.
References
-
- Harder J. D. Matses Indian rainforest habitat classification and mammalian diversity in Amazonian Peru. J. Ethnobiol. 20, 1–36 (2000) .
-
- Israel B. A., Schulz A. J., Parker E. A. & Becker A. B. Review of community-based research: assessing partnership approaches to improve public health. Annu. Rev. Public Health 19, 173–202 (1998) . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

