RNA chaperones buffer deleterious mutations in E. coli
- PMID: 25806682
- PMCID: PMC4402597
- DOI: 10.7554/eLife.04745
RNA chaperones buffer deleterious mutations in E. coli
Abstract
Both proteins and RNAs can misfold into non-functional conformations. Protein chaperones promote native folding of nascent polypeptides and refolding of misfolded species, thereby buffering mutations that compromise protein structure and function. Here, we show that RNA chaperones can also act as mutation buffers that enhance organismal fitness. Using competition assays, we demonstrate that overexpression of select RNA chaperones, including three DEAD box RNA helicases (DBRHs) (CsdA, SrmB, RhlB) and the cold shock protein CspA, improves fitness of two independently evolved Escherichia coli mutator strains that have accumulated deleterious mutations during short- and long-term laboratory evolution. We identify strain-specific mutations that are deleterious and subject to buffering when introduced individually into the ancestral genotype. For DBRHs, we show that buffering requires helicase activity, implicating RNA structural remodelling in the buffering process. Our results suggest that RNA chaperones might play a fundamental role in RNA evolution and evolvability.
Keywords: E. coli; chaperones; evolution; evolutionary biology; genomics; mutation buffering.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
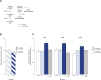
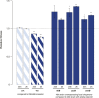
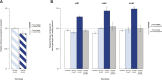
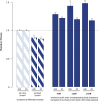

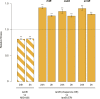
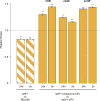
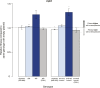
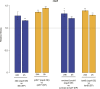

Similar articles
-
Allosteric activation of the ATPase activity of the Escherichia coli RhlB RNA helicase.J Biol Chem. 2008 Feb 29;283(9):5567-76. doi: 10.1074/jbc.M708620200. Epub 2007 Dec 28. J Biol Chem. 2008. PMID: 18165229 Free PMC article.
-
CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit.Nucleic Acids Res. 2004 May 17;32(9):2751-9. doi: 10.1093/nar/gkh603. Print 2004. Nucleic Acids Res. 2004. PMID: 15148362 Free PMC article.
-
Identification of the sites of action of SrmB, a DEAD-box RNA helicase involved in Escherichia coli ribosome assembly.Mol Microbiol. 2011 Oct;82(2):300-11. doi: 10.1111/j.1365-2958.2011.07779.x. Epub 2011 Aug 22. Mol Microbiol. 2011. PMID: 21859437
-
Assembly of bacterial ribosomes.Annu Rev Biochem. 2011;80:501-26. doi: 10.1146/annurev-biochem-062608-160432. Annu Rev Biochem. 2011. PMID: 21529161 Review.
-
Proteins That Chaperone RNA Regulation.Microbiol Spectr. 2018 Jul;6(4):10.1128/microbiolspec.rwr-0026-2018. doi: 10.1128/microbiolspec.RWR-0026-2018. Microbiol Spectr. 2018. PMID: 30051798 Free PMC article. Review.
Cited by
-
FinO/ProQ-family proteins: an evolutionary perspective.Biosci Rep. 2023 Mar 31;43(3):BSR20220313. doi: 10.1042/BSR20220313. Biosci Rep. 2023. PMID: 36787218 Free PMC article. Review.
-
Precision Genome Engineering in Streptococcus suis Based on a Broad-Host-Range Vector and CRISPR-Cas9 Technology.ACS Synth Biol. 2023 Sep 15;12(9):2546-2560. doi: 10.1021/acssynbio.3c00110. Epub 2023 Aug 21. ACS Synth Biol. 2023. PMID: 37602730 Free PMC article.
-
A 2D-DIGE-based proteomic analysis brings new insights into cellular responses of Pseudomonas putida KT2440 during polyhydroxyalkanoates synthesis.Microb Cell Fact. 2019 May 28;18(1):93. doi: 10.1186/s12934-019-1146-5. Microb Cell Fact. 2019. PMID: 31138236 Free PMC article.
-
Isolating Escherichia coli strains for recombinant protein production.Cell Mol Life Sci. 2017 Mar;74(5):891-908. doi: 10.1007/s00018-016-2371-2. Epub 2016 Oct 11. Cell Mol Life Sci. 2017. PMID: 27730255 Free PMC article. Review.
-
Cold Shock Proteins: A Minireview with Special Emphasis on Csp-family of Enteropathogenic Yersinia.Front Microbiol. 2016 Jul 22;7:1151. doi: 10.3389/fmicb.2016.01151. eCollection 2016. Front Microbiol. 2016. PMID: 27499753 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

