Global variability in gene expression and alternative splicing is modulated by mitochondrial content
- PMID: 25800673
- PMCID: PMC4417112
- DOI: 10.1101/gr.178426.114
Global variability in gene expression and alternative splicing is modulated by mitochondrial content
Abstract
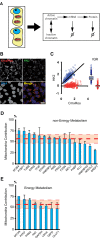
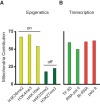
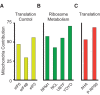
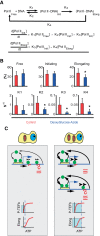
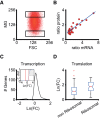
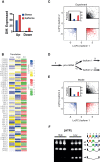
Noise in gene expression is a main determinant of phenotypic variability. Increasing experimental evidence suggests that genome-wide cellular constraints largely contribute to the heterogeneity observed in gene products. It is still unclear, however, which global factors affect gene expression noise and to what extent. Since eukaryotic gene expression is an energy demanding process, differences in the energy budget of each cell could determine gene expression differences. Here, we quantify the contribution of mitochondrial variability (a natural source of ATP variation) to global variability in gene expression. We find that changes in mitochondrial content can account for ∼50% of the variability observed in protein levels. This is the combined result of the effect of mitochondria dosage on transcription and translation apparatus content and activities. Moreover, we find that mitochondrial levels have a large impact on alternative splicing, thus modulating both the abundance and type of mRNAs. A simple mathematical model in which mitochondrial content simultaneously affects transcription rate and splicing site choice can explain the alternative splicing data. The results of this study show that mitochondrial content (and/or probably function) influences mRNA abundance, translation, and alternative splicing, which ultimately affects cellular phenotype.
© 2015 Guantes et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
Similar articles
-
Mitochondria and the non-genetic origins of cell-to-cell variability: More is different.Bioessays. 2016 Jan;38(1):64-76. doi: 10.1002/bies.201500082. Epub 2015 Dec 12. Bioessays. 2016. PMID: 26660201 Review.
-
Regulation of the cell cycle via mitochondrial gene expression and energy metabolism in HeLa cells.Acta Biochim Biophys Sin (Shanghai). 2012 Apr;44(4):347-58. doi: 10.1093/abbs/gms006. Epub 2012 Feb 16. Acta Biochim Biophys Sin (Shanghai). 2012. PMID: 22343378
-
Mitochondrial damage modulates alternative splicing in neuronal cells: implications for neurodegeneration.J Neurochem. 2007 Jan;100(1):142-53. doi: 10.1111/j.1471-4159.2006.04204.x. Epub 2006 Oct 25. J Neurochem. 2007. PMID: 17064354
-
Nuclear and mitochondrial genome responses in HeLa cells treated with inhibitors of mitochondrial DNA expression.Acta Biochim Pol. 2006;53(3):485-95. Epub 2006 Sep 2. Acta Biochim Pol. 2006. PMID: 16951738
-
Mitochondrial topoisomerases and alternative splicing of the human TOP1mt gene.Biochimie. 2007 Apr;89(4):474-81. doi: 10.1016/j.biochi.2006.11.002. Epub 2006 Nov 27. Biochimie. 2007. PMID: 17161897 Review.
Cited by
-
Mitochondrial DNA copy number can influence mortality and cardiovascular disease via methylation of nuclear DNA CpGs.Genome Med. 2020 Sep 28;12(1):84. doi: 10.1186/s13073-020-00778-7. Genome Med. 2020. PMID: 32988399 Free PMC article.
-
What's Luck Got to Do with It: Single Cells, Multiple Fates, and Biological Nondeterminism.Mol Cell. 2016 Jun 2;62(5):788-802. doi: 10.1016/j.molcel.2016.05.023. Mol Cell. 2016. PMID: 27259209 Free PMC article. Review.
-
Mitochondrial DNA copy number reduction via in vitro TFAM knockout remodels the nuclear epigenome and transcriptome.bioRxiv [Preprint]. 2024 Feb 10:2024.01.29.577835. doi: 10.1101/2024.01.29.577835. bioRxiv. 2024. PMID: 38352513 Free PMC article. Preprint.
-
Increased gene expression noise in human cancers is correlated with low p53 and immune activities as well as late stage cancer.Oncotarget. 2016 Nov 1;7(44):72011-72020. doi: 10.18632/oncotarget.12457. Oncotarget. 2016. PMID: 27713130 Free PMC article.
-
A model to predict a risk of allergic rhinitis based on mitochondrial DNA copy number.Eur Arch Otorhinolaryngol. 2022 Oct;279(10):4997-5008. doi: 10.1007/s00405-022-07341-7. Epub 2022 Mar 29. Eur Arch Otorhinolaryngol. 2022. PMID: 35348857
References
-
- Brock A, Chang H, Huang S. 2009. Non-genetic heterogeneity—a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet 10: 336–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
