N6-methyladenosine marks primary microRNAs for processing
- PMID: 25799998
- PMCID: PMC4475635
- DOI: 10.1038/nature14281
N6-methyladenosine marks primary microRNAs for processing
Abstract
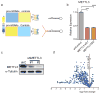
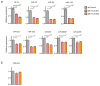
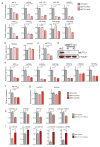
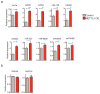
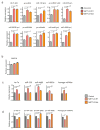
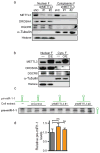
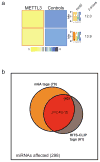
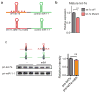
The first step in the biogenesis of microRNAs is the processing of primary microRNAs (pri-miRNAs) by the microprocessor complex, composed of the RNA-binding protein DGCR8 and the type III RNase DROSHA. This initial event requires recognition of the junction between the stem and the flanking single-stranded RNA of the pri-miRNA hairpin by DGCR8 followed by recruitment of DROSHA, which cleaves the RNA duplex to yield the pre-miRNA product. While the mechanisms underlying pri-miRNA processing have been determined, the mechanism by which DGCR8 recognizes and binds pri-miRNAs, as opposed to other secondary structures present in transcripts, is not understood. Here we find in mammalian cells that methyltransferase-like 3 (METTL3) methylates pri-miRNAs, marking them for recognition and processing by DGCR8. Consistent with this, METTL3 depletion reduced the binding of DGCR8 to pri-miRNAs and resulted in the global reduction of mature miRNAs and concomitant accumulation of unprocessed pri-miRNAs. In vitro processing reactions confirmed the sufficiency of the N(6)-methyladenosine (m(6)A) mark in promoting pri-miRNA processing. Finally, gain-of-function experiments revealed that METTL3 is sufficient to enhance miRNA maturation in a global and non-cell-type-specific manner. Our findings reveal that the m(6)A mark acts as a key post-transcriptional modification that promotes the initiation of miRNA biogenesis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
The core microprocessor component DiGeorge syndrome critical region 8 (DGCR8) is a nonspecific RNA-binding protein.J Biol Chem. 2013 Sep 13;288(37):26785-99. doi: 10.1074/jbc.M112.446880. Epub 2013 Jul 26. J Biol Chem. 2013. PMID: 23893406 Free PMC article.
-
Bulges control pri-miRNA processing in a position and strand-dependent manner.RNA Biol. 2021 Nov;18(11):1716-1726. doi: 10.1080/15476286.2020.1868139. Epub 2020 Dec 31. RNA Biol. 2021. PMID: 33382955 Free PMC article.
-
A central role for the primary microRNA stem in guiding the position and efficiency of Drosha processing of a viral pri-miRNA.RNA. 2014 Jul;20(7):1068-77. doi: 10.1261/rna.044537.114. Epub 2014 May 22. RNA. 2014. PMID: 24854622 Free PMC article.
-
Autoregulatory mechanisms controlling the Microprocessor.Adv Exp Med Biol. 2010;700:56-66. Adv Exp Med Biol. 2010. PMID: 21627030 Review.
-
MicroRNA biogenesis: isolation and characterization of the microprocessor complex.Methods Mol Biol. 2006;342:33-47. doi: 10.1385/1-59745-123-1:33. Methods Mol Biol. 2006. PMID: 16957365 Review.
Cited by
-
Reshaping the role of m6A modification in cancer transcriptome: a review.Cancer Cell Int. 2020 Jul 29;20:353. doi: 10.1186/s12935-020-01445-y. eCollection 2020. Cancer Cell Int. 2020. PMID: 32760220 Free PMC article. Review.
-
Association between METTL3 gene polymorphisms and neuroblastoma susceptibility: A nine-centre case-control study.J Cell Mol Med. 2020 Aug;24(16):9280-9286. doi: 10.1111/jcmm.15576. Epub 2020 Jul 2. J Cell Mol Med. 2020. PMID: 32615646 Free PMC article.
-
Crosstalk between RNA m6A Modification and Non-coding RNA Contributes to Cancer Growth and Progression.Mol Ther Nucleic Acids. 2020 Aug 8;22:62-71. doi: 10.1016/j.omtn.2020.08.004. Online ahead of print. Mol Ther Nucleic Acids. 2020. PMID: 32911345 Free PMC article. Review.
-
MicroRNAs in cardiovascular ageing.J Physiol. 2016 Apr 15;594(8):2085-94. doi: 10.1113/JP270557. Epub 2015 Jul 5. J Physiol. 2016. PMID: 26040259 Free PMC article. Review.
-
Exploration of N6-Methyladenosine Profiles of mRNAs and the Function of METTL3 in Atherosclerosis.Cells. 2022 Sep 24;11(19):2980. doi: 10.3390/cells11192980. Cells. 2022. PMID: 36230944 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

