Ferroptosis as a p53-mediated activity during tumour suppression
- PMID: 25799988
- PMCID: PMC4455927
- DOI: 10.1038/nature14344
Ferroptosis as a p53-mediated activity during tumour suppression
Abstract
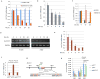
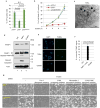
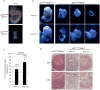
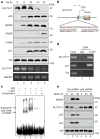
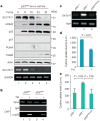
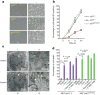
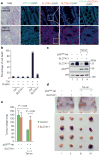
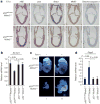
Although p53-mediated cell-cycle arrest, senescence and apoptosis serve as critical barriers to cancer development, emerging evidence suggests that the metabolic activities of p53 are also important. Here we show that p53 inhibits cystine uptake and sensitizes cells to ferroptosis, a non-apoptotic form of cell death, by repressing expression of SLC7A11, a key component of the cystine/glutamate antiporter. Notably, p53(3KR), an acetylation-defective mutant that fails to induce cell-cycle arrest, senescence and apoptosis, fully retains the ability to regulate SLC7A11 expression and induce ferroptosis upon reactive oxygen species (ROS)-induced stress. Analysis of mutant mice shows that these non-canonical p53 activities contribute to embryonic development and the lethality associated with loss of Mdm2. Moreover, SLC7A11 is highly expressed in human tumours, and its overexpression inhibits ROS-induced ferroptosis and abrogates p53(3KR)-mediated tumour growth suppression in xenograft models. Our findings uncover a new mode of tumour suppression based on p53 regulation of cystine metabolism, ROS responses and ferroptosis.
Figures






Comment in
-
Cancer: A piece of the p53 puzzle.Nature. 2015 Apr 2;520(7545):37-8. doi: 10.1038/nature14374. Epub 2015 Mar 18. Nature. 2015. PMID: 25799989 No abstract available.
Similar articles
-
Dynamic roles of p53-mediated metabolic activities in ROS-induced stress responses.Cell Cycle. 2015;14(18):2881-5. doi: 10.1080/15384101.2015.1068479. Cell Cycle. 2015. PMID: 26218928 Free PMC article.
-
Acetylation Is Crucial for p53-Mediated Ferroptosis and Tumor Suppression.Cell Rep. 2016 Oct 4;17(2):366-373. doi: 10.1016/j.celrep.2016.09.022. Cell Rep. 2016. PMID: 27705786 Free PMC article.
-
Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses.Proc Natl Acad Sci U S A. 2016 Nov 1;113(44):E6806-E6812. doi: 10.1073/pnas.1607152113. Epub 2016 Oct 3. Proc Natl Acad Sci U S A. 2016. PMID: 27698118 Free PMC article.
-
Cystine-glutamate antiporter xCT as a therapeutic target for cancer.Cell Biochem Funct. 2021 Mar;39(2):174-179. doi: 10.1002/cbf.3581. Epub 2020 Aug 4. Cell Biochem Funct. 2021. PMID: 32749001 Review.
-
Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy.Protein Cell. 2021 Aug;12(8):599-620. doi: 10.1007/s13238-020-00789-5. Epub 2020 Oct 1. Protein Cell. 2021. PMID: 33000412 Free PMC article. Review.
Cited by
-
SLC7A9 suppression increases chemosensitivity by inducing ferroptosis via the inhibition of cystine transport in gastric cancer.EBioMedicine. 2024 Nov;109:105375. doi: 10.1016/j.ebiom.2024.105375. Epub 2024 Oct 21. EBioMedicine. 2024. PMID: 39437660 Free PMC article.
-
Unlocking p53 response elements: DNA shape is the key.Mol Cell Oncol. 2021 Apr 22;8(3):1905489. doi: 10.1080/23723556.2021.1905489. eCollection 2021. Mol Cell Oncol. 2021. PMID: 34027043 Free PMC article.
-
The Impacts of Iron Overload and Ferroptosis on Intestinal Mucosal Homeostasis and Inflammation.Int J Mol Sci. 2022 Nov 17;23(22):14195. doi: 10.3390/ijms232214195. Int J Mol Sci. 2022. PMID: 36430673 Free PMC article. Review.
-
Ferroptosis in head and neck squamous cell carcinoma: from pathogenesis to treatment.Front Pharmacol. 2024 Jan 19;15:1283465. doi: 10.3389/fphar.2024.1283465. eCollection 2024. Front Pharmacol. 2024. PMID: 38313306 Free PMC article. Review.
-
Identification of Ferroptosis-Related Molecular Clusters and Immune Characterization in Autism Spectrum Disorder.Front Genet. 2022 Aug 11;13:911119. doi: 10.3389/fgene.2022.911119. eCollection 2022. Front Genet. 2022. PMID: 36035135 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- P01 CA097403/CA/NCI NIH HHS/United States
- 2P01CA080058/CA/NCI NIH HHS/United States
- R01 CA172272/CA/NCI NIH HHS/United States
- R01 CA166294/CA/NCI NIH HHS/United States
- 5R01CA172023/CA/NCI NIH HHS/United States
- P01 CA080058/CA/NCI NIH HHS/United States
- 2P01CA097403/CA/NCI NIH HHS/United States
- R01 CA172023/CA/NCI NIH HHS/United States
- 5R01CA169246/CA/NCI NIH HHS/United States
- 5R01CA166294/CA/NCI NIH HHS/United States
- R01 CA169246/CA/NCI NIH HHS/United States
- R01 CA085533/CA/NCI NIH HHS/United States
- T32-CA09503/CA/NCI NIH HHS/United States
- T32 CA009503/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

