The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection
- PMID: 25774715
- PMCID: PMC4406811
- DOI: 10.1038/ni.3118
The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection
Abstract
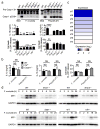
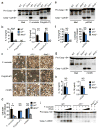
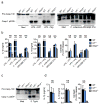
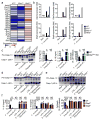
Inflammasomes are critical for mounting host defense against pathogens. The molecular mechanisms that control activation of the AIM2 inflammasome in response to different cytosolic pathogens remain unclear. Here we found that the transcription factor IRF1 was required for activation of the AIM2 inflammasome during infection with the Francisella tularensis subspecies novicida (F. novicida), whereas engagement of the AIM2 inflammasome by mouse cytomegalovirus (MCMV) or transfected double-stranded DNA did not require IRF1. Infection of F. novicida detected by the DNA sensor cGAS and its adaptor STING induced type I interferon-dependent expression of IRF1, which drove the expression of guanylate-binding proteins (GBPs); this led to intracellular killing of bacteria and DNA release. Our results reveal a specific requirement for IRF1 and GBPs in the liberation of DNA for sensing by AIM2 depending on the pathogen encountered by the cell.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida.Nat Immunol. 2015 May;16(5):476-484. doi: 10.1038/ni.3119. Epub 2015 Mar 16. Nat Immunol. 2015. PMID: 25774716 Free PMC article.
-
GBPs take AIM at Francisella.Nat Immunol. 2015 May;16(5):443-4. doi: 10.1038/ni.3144. Nat Immunol. 2015. PMID: 25898190 No abstract available.
-
IFN-γ extends the immune functions of Guanylate Binding Proteins to inflammasome-independent antibacterial activities during Francisella novicida infection.PLoS Pathog. 2017 Oct 2;13(10):e1006630. doi: 10.1371/journal.ppat.1006630. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 28968459 Free PMC article.
-
Francisella Inflammasomes: Integrated Responses to a Cytosolic Stealth Bacterium.Curr Top Microbiol Immunol. 2016;397:229-56. doi: 10.1007/978-3-319-41171-2_12. Curr Top Microbiol Immunol. 2016. PMID: 27460813 Review.
-
Interferon-inducible guanylate-binding proteins at the interface of cell-autonomous immunity and inflammasome activation.J Leukoc Biol. 2017 Jan;101(1):143-150. doi: 10.1189/jlb.4MR0516-223R. Epub 2016 Jul 14. J Leukoc Biol. 2017. PMID: 27418355 Free PMC article. Review.
Cited by
-
Inflammasomes: mechanism of action, role in disease, and therapeutics.Nat Med. 2015 Jul;21(7):677-87. doi: 10.1038/nm.3893. Epub 2015 Jun 29. Nat Med. 2015. PMID: 26121197 Free PMC article. Review.
-
AIM2 in health and disease: Inflammasome and beyond.Immunol Rev. 2020 Sep;297(1):83-95. doi: 10.1111/imr.12903. Epub 2020 Jul 26. Immunol Rev. 2020. PMID: 32713036 Free PMC article. Review.
-
Human GBP1 Differentially Targets Salmonella and Toxoplasma to License Recognition of Microbial Ligands and Caspase-Mediated Death.Cell Rep. 2020 Aug 11;32(6):108008. doi: 10.1016/j.celrep.2020.108008. Cell Rep. 2020. PMID: 32783936 Free PMC article.
-
The multiple roles of interferon regulatory factor family in health and disease.Signal Transduct Target Ther. 2024 Oct 9;9(1):282. doi: 10.1038/s41392-024-01980-4. Signal Transduct Target Ther. 2024. PMID: 39384770 Free PMC article. Review.
-
Intravesical BCG in patients with non-muscle invasive bladder cancer induces trained immunity and decreases respiratory infections.J Immunother Cancer. 2023 Jan;11(1):e005518. doi: 10.1136/jitc-2022-005518. J Immunother Cancer. 2023. PMID: 36693678 Free PMC article.
References
-
- Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481(7381):278–286. - PubMed
-
- Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–1022. - PubMed
-
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature immunology. 2009;10(3):266–272. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

