Long non-coding RNAs control hematopoietic stem cell function
- PMID: 25772072
- PMCID: PMC4388783
- DOI: 10.1016/j.stem.2015.02.002
Long non-coding RNAs control hematopoietic stem cell function
Abstract
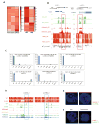
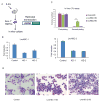
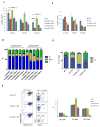
Hematopoietic stem cells (HSCs) possess unique gene expression programs that enforce their identity and regulate lineage commitment. Long non-coding RNAs (lncRNAs) have emerged as important regulators of gene expression and cell fate decisions, although their functions in HSCs are unclear. Here we profiled the transcriptome of purified HSCs by deep sequencing and identified 323 unannotated lncRNAs. Comparing their expression in differentiated lineages revealed 159 lncRNAs enriched in HSCs, some of which are likely HSC specific (LncHSCs). These lncRNA genes share epigenetic features with protein-coding genes, including regulated expression via DNA methylation, and knocking down two LncHSCs revealed distinct effects on HSC self-renewal and lineage commitment. We mapped the genomic binding sites of one of these candidates and found enrichment for key hematopoietic transcription factor binding sites, especially E2A. Together, these results demonstrate that lncRNAs play important roles in regulating HSCs, providing an additional layer to the genetic circuitry controlling HSC function.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Long noncoding RNAs of single hematopoietic stem and progenitor cells in healthy and dysplastic human bone marrow.Haematologica. 2019 May;104(5):894-906. doi: 10.3324/haematol.2018.208926. Epub 2018 Dec 13. Haematologica. 2019. PMID: 30545929 Free PMC article.
-
Combined Single-Cell Profiling of lncRNAs and Functional Screening Reveals that H19 Is Pivotal for Embryonic Hematopoietic Stem Cell Development.Cell Stem Cell. 2019 Feb 7;24(2):285-298.e5. doi: 10.1016/j.stem.2018.11.023. Epub 2019 Jan 10. Cell Stem Cell. 2019. PMID: 30639035
-
Deciphering Transcriptomic Variations in Hematopoietic Lineages: HSCs, EBs, and MKs.Int J Mol Sci. 2024 Sep 19;25(18):10073. doi: 10.3390/ijms251810073. Int J Mol Sci. 2024. PMID: 39337559 Free PMC article.
-
Long non-coding RNA: Classification, biogenesis and functions in blood cells.Mol Immunol. 2019 Aug;112:82-92. doi: 10.1016/j.molimm.2019.04.011. Epub 2019 May 9. Mol Immunol. 2019. PMID: 31079005 Review.
-
Altering chromatin methylation patterns and the transcriptional network involved in regulation of hematopoietic stem cell fate.J Cell Physiol. 2020 Oct;235(10):6404-6423. doi: 10.1002/jcp.29642. Epub 2020 Feb 13. J Cell Physiol. 2020. PMID: 32052445 Review.
Cited by
-
The Mechanism of Stem Cell Aging.Stem Cell Rev Rep. 2022 Apr;18(4):1281-1293. doi: 10.1007/s12015-021-10317-5. Epub 2022 Jan 9. Stem Cell Rev Rep. 2022. PMID: 35000109 Free PMC article. Review.
-
Hematopoietic Stem Cells in Wound Healing Response.Adv Wound Care (New Rochelle). 2022 Nov;11(11):598-621. doi: 10.1089/wound.2021.0065. Epub 2021 Sep 9. Adv Wound Care (New Rochelle). 2022. PMID: 34353116 Free PMC article.
-
Extracellular vesicles of stromal origin target and support hematopoietic stem and progenitor cells.J Cell Biol. 2017 Jul 3;216(7):2217-2230. doi: 10.1083/jcb.201601109. Epub 2017 Jun 19. J Cell Biol. 2017. PMID: 28630143 Free PMC article.
-
Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis.Blood. 2017 Nov 2;130(18):1965-1975. doi: 10.1182/blood-2017-06-788695. Epub 2017 Sep 19. Blood. 2017. PMID: 28928124 Free PMC article. Review.
-
Long non-coding RNAs and MYC association in hematological malignancies.Ann Hematol. 2020 Oct;99(10):2231-2242. doi: 10.1007/s00277-020-04166-4. Epub 2020 Jul 4. Ann Hematol. 2020. PMID: 32621182 Review.
References
-
- Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, Weichenhan D, Lier A, von Paleske L, Renders S, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA Methylome analysis. Cell Stem Cell. 2014;15:507–522. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
- CA125123/CA/NCI NIH HHS/United States
- HD007495/HD/NICHD NIH HHS/United States
- K99 DK084259/DK/NIDDK NIH HHS/United States
- R56 DK092883/DK/NIDDK NIH HHS/United States
- DK092883/DK/NIDDK NIH HHS/United States
- AI036211/AI/NIAID NIH HHS/United States
- T32 HD007495/HD/NICHD NIH HHS/United States
- AG036562/AG/NIA NIH HHS/United States
- R01HG007538/HG/NHGRI NIH HHS/United States
- DK084259/DK/NIDDK NIH HHS/United States
- T32 AI007495/AI/NIAID NIH HHS/United States
- P50 CA126752/CA/NCI NIH HHS/United States
- U54 HD007495/HD/NICHD NIH HHS/United States
- P30 CA125123/CA/NCI NIH HHS/United States
- P30 DK056338/DK/NIDDK NIH HHS/United States
- 5T32AI007495/AI/NIAID NIH HHS/United States
- DK56338/DK/NIDDK NIH HHS/United States
- P30 AI036211/AI/NIAID NIH HHS/United States
- CA126752/CA/NCI NIH HHS/United States
- S10 RR024574/RR/NCRR NIH HHS/United States
- RC2 AG036562/AG/NIA NIH HHS/United States
- T32 DK060445/DK/NIDDK NIH HHS/United States
- R01 HG007538/HG/NHGRI NIH HHS/United States
- R00 DK084259/DK/NIDDK NIH HHS/United States
- AG28865/AG/NIA NIH HHS/United States
- R01 DK092883/DK/NIDDK NIH HHS/United States
- P30 HD007495/HD/NICHD NIH HHS/United States
- RR024574/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

