Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease
- PMID: 25764541
- PMCID: PMC4358749
- DOI: 10.1093/femsec/fiu002
Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease
Abstract
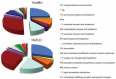
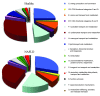
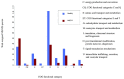
Obesity is becoming the new pediatric epidemic. Non-alcoholic fatty liver disease (NAFLD) is frequently associated with obesity and has become the most common cause of pediatric liver disease. The gut microbiome is the major metabolic organ and determines how calories are processed, serving as a caloric gate and contributing towards the pathogenesis of NAFLD. The goal of this study is to examine gut microbial profiles in children with NAFLD using phylogenetic, metabolomic, metagenomic and proteomic approaches. Fecal samples were obtained from obese children with or without NAFLD and healthy lean children. Stool specimens were subjected to 16S rRNA gene microarray, shotgun sequencing, mass spectroscopy for proteomics and NMR spectroscopy for metabolite analysis. Children with NAFLD had more abundant Gammaproteobacteria and Prevotella and significantly higher levels of ethanol, with differential effects on short chain fatty acids. This group also had increased genomic and protein abundance for energy production with a reduction in carbohydrate and amino acid metabolism and urea cycle and urea transport systems. The metaproteome and metagenome showed similar findings. The gut microbiome in pediatric NAFLD is distinct from lean healthy children with more alcohol production and pathways allocated to energy metabolism over carbohydrate and amino acid metabolism, which would contribute to development of disease.
Keywords: NASH; carbohydrate; energy metabolism; gut microbiome; obesity.
© FEMS 2014. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
Microbiome Signatures Associated With Steatohepatitis and Moderate to Severe Fibrosis in Children With Nonalcoholic Fatty Liver Disease.Gastroenterology. 2019 Oct;157(4):1109-1122. doi: 10.1053/j.gastro.2019.06.028. Epub 2019 Jun 27. Gastroenterology. 2019. PMID: 31255652 Free PMC article.
-
Role of intestinal flora in the development of nonalcoholic fatty liver disease in children.Microbiol Spectr. 2024 Feb 6;12(2):e0100623. doi: 10.1128/spectrum.01006-23. Epub 2024 Jan 8. Microbiol Spectr. 2024. PMID: 38189294 Free PMC article.
-
A metagenomic study identifies a Prevotella copri enriched microbial profile associated with non-alcoholic steatohepatitis in subjects with obesity.J Gastroenterol Hepatol. 2023 May;38(5):791-799. doi: 10.1111/jgh.16147. Epub 2023 Mar 2. J Gastroenterol Hepatol. 2023. PMID: 36807933
-
Understanding the Role of the Gut Microbiome and Microbial Metabolites in Non-Alcoholic Fatty Liver Disease: Current Evidence and Perspectives.Biomolecules. 2021 Dec 31;12(1):56. doi: 10.3390/biom12010056. Biomolecules. 2021. PMID: 35053205 Free PMC article. Review.
-
Role of gut microbiota and Toll-like receptors in nonalcoholic fatty liver disease.World J Gastroenterol. 2014 Jun 21;20(23):7381-91. doi: 10.3748/wjg.v20.i23.7381. World J Gastroenterol. 2014. PMID: 24966608 Free PMC article. Review.
Cited by
-
Exercise training modulates the gut microbiota profile and impairs inflammatory signaling pathways in obese children.Exp Mol Med. 2020 Jul;52(7):1048-1061. doi: 10.1038/s12276-020-0459-0. Epub 2020 Jul 6. Exp Mol Med. 2020. PMID: 32624568 Free PMC article. Clinical Trial.
-
Prebiotic Xylo-Oligosaccharides Ameliorate High-Fat-Diet-Induced Hepatic Steatosis in Rats.Nutrients. 2020 Oct 22;12(11):3225. doi: 10.3390/nu12113225. Nutrients. 2020. PMID: 33105554 Free PMC article.
-
Fecal Metagenomics and Metabolomics Identifying Microbial Signatures in Non-Alcoholic Fatty Liver Disease.Int J Mol Sci. 2023 Mar 2;24(5):4855. doi: 10.3390/ijms24054855. Int J Mol Sci. 2023. PMID: 36902288 Free PMC article. Review.
-
Microbial metabolites in non-alcoholic fatty liver disease.World J Gastroenterol. 2019 May 7;25(17):2019-2028. doi: 10.3748/wjg.v25.i17.2019. World J Gastroenterol. 2019. PMID: 31114130 Free PMC article. Review.
-
Faecalibacterium prausnitzii prevents hepatic damage in a mouse model of NASH induced by a high-fructose high-fat diet.Front Microbiol. 2023 Mar 16;14:1123547. doi: 10.3389/fmicb.2023.1123547. eCollection 2023. Front Microbiol. 2023. PMID: 37007480 Free PMC article.
References
-
- Anderson PE, Raymer ML, Kelly BJ, et al. Characterization of 1H NMR spectroscopic data and the generation of synthetic validation sets. Bioinformatics. 2009;25:2992–3000. - PubMed
-
- Anderson PEM, Mahle DA, Doom TE, et al. Dynamic adaptive binning: an improved quantification technique for NMR spectroscopic data. Metabolomics. 2011;7:179–90.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

