Dynamics of Human Cytomegalovirus Infection in CD34+ Hematopoietic Cells and Derived Langerhans-Type Dendritic Cells
- PMID: 25762731
- PMCID: PMC4442541
- DOI: 10.1128/JVI.00305-15
Dynamics of Human Cytomegalovirus Infection in CD34+ Hematopoietic Cells and Derived Langerhans-Type Dendritic Cells
Abstract
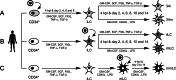
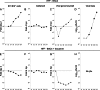
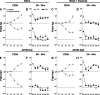
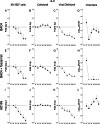
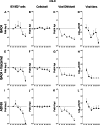
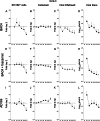
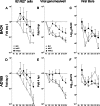
Acquisition of human cytomegalovirus (CMV) usually occurs by contact between contaminated bodily fluids, such as urine and saliva, and host mucosal cells. Langerhans-type dendritic cells (LC) are the only type of immune cells found in the outermost layers of the oral mucosae, where they not only provide a first line of defense against CMV but can easily be targeted by orally administered vaccines, while their bone marrow resident progenitors are important sites of virus latency. In this work, we tracked the progress of infection in CD34(+) progenitor cells, immature LC (iLC), and mature LC (mLC) exposed to the clinical-like strain TB40-BAC4 or to the vaccine strain AD169varATCC, prior to their long-term maintenance under either immature or mature conditions. We show that the genomes of both strains are efficiently maintained in CD34(+) cells during their differentiation into iLC, although this requires the presence of larger amounts of input AD169varATCC DNA. Lipopolysaccharide- and CD40 ligand-induced maturation of iLC derived from latently infected progenitors was not associated with robust viral genome replication and progeny production, while maturation of directly infected iLC increased and prolonged expression of the viral immediate early proteins. While effective replication of viral genomes from both strains occurred only in mLC, both iLC and mLC produced viral progeny, suggesting that both types of LC may contribute to CMV horizontal transmission in vivo.
Importance: Human CMV is usually acquired via the oral and nasal mucosae. Langerhans-type dendritic cells (LC) are the only type of immune cells found in the outermost layers of these tissues. Understanding how CMV interacts with LC and their hematopoietic progenitors is thus essential to develop innovative means of defense against this virus. Here we show that the genomes of a virulent and an attenuated strain of CMV are maintained in hematopoietic progenitor cells during their differentiation into immature LC and that maturation of these cells by exposure to lipopolysaccharide and CD40 ligand is not sufficient to trigger virus reactivation. While the extents of viral protein expression and genome replication were broadest in directly infected mature LC populations, similar amounts of viral progeny were detected in the supernatants of immature and mature LC, suggesting that these immune cells of the oral mucosa are likely to be important for CMV transmission within the human population.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Susceptibility of immature and mature Langerhans cell-type dendritic cells to infection and immunomodulation by human cytomegalovirus.J Virol. 2003 Jul;77(13):7563-74. doi: 10.1128/jvi.77.13.7563-7574.2003. J Virol. 2003. PMID: 12805456 Free PMC article.
-
Human Cytomegalovirus US28 Ligand Binding Activity Is Required for Latency in CD34+ Hematopoietic Progenitor Cells and Humanized NSG Mice.mBio. 2019 Aug 20;10(4):e01889-19. doi: 10.1128/mBio.01889-19. mBio. 2019. PMID: 31431555 Free PMC article.
-
Human cytomegalovirus infection of langerhans-type dendritic cells does not require the presence of the gH/gL/UL128-131A complex and is blocked after nuclear deposition of viral genomes in immature cells.J Virol. 2014 Jan;88(1):403-16. doi: 10.1128/JVI.03062-13. Epub 2013 Oct 23. J Virol. 2014. PMID: 24155395 Free PMC article.
-
Cytomegalovirus latency and latency-specific transcription in hematopoietic progenitors.Scand J Infect Dis Suppl. 1995;99:63-7. Scand J Infect Dis Suppl. 1995. PMID: 8668945 Review.
-
Human cytomegalovirus tropism for mucosal myeloid dendritic cells.Rev Med Virol. 2014 Nov;24(6):379-95. doi: 10.1002/rmv.1797. Epub 2014 Jun 2. Rev Med Virol. 2014. PMID: 24888709 Free PMC article. Review.
Cited by
-
Latent human cytomegalovirus enhances HIV-1 infection in CD34+ progenitor cells.Blood Adv. 2017 Jan 16;1(5):306-318. doi: 10.1182/bloodadvances.2016000638. eCollection 2017 Jan 24. Blood Adv. 2017. PMID: 29296946 Free PMC article.
-
New Insights Into the Molecular Mechanisms and Immune Control of Cytomegalovirus Reactivation.Transplantation. 2020 May;104(5):e118-e124. doi: 10.1097/TP.0000000000003138. Transplantation. 2020. PMID: 31996662 Free PMC article. Review.
-
Human cytomegalovirus G protein-coupled receptor US28 promotes latency by attenuating c-fos.Proc Natl Acad Sci U S A. 2019 Jan 29;116(5):1755-1764. doi: 10.1073/pnas.1816933116. Epub 2019 Jan 15. Proc Natl Acad Sci U S A. 2019. PMID: 30647114 Free PMC article.
-
Cytomegalovirus Strain TB40/E Restrictions and Adaptations to Growth in ARPE-19 Epithelial Cells.Microorganisms. 2020 Apr 24;8(4):615. doi: 10.3390/microorganisms8040615. Microorganisms. 2020. PMID: 32344555 Free PMC article.
-
Human Cytomegalovirus Latency: Approaching the Gordian Knot.Annu Rev Virol. 2016 Sep 29;3(1):333-357. doi: 10.1146/annurev-virology-110615-042422. Epub 2016 Aug 4. Annu Rev Virol. 2016. PMID: 27501258 Free PMC article.
References
-
- Zhuravskaya T, Maciejewski JP, Netski DM, Bruening E, Mackintosh FR, St Jeor S. 1997. Spread of human cytomegalovirus (HCMV) after infection of human hematopoietic progenitor cells: model of HCMV latency. Blood 90:2482–2491. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

