Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing
- PMID: 25759497
- PMCID: PMC4453535
- DOI: 10.1128/mBio.00054-15
Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing
Abstract
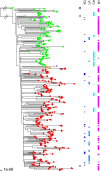
Methicillin-resistant Staphylococcus aureus (MRSA) USA300 is a successful S. aureus clone in the United States and a common cause of skin and soft tissue infections (SSTIs). We performed whole-genome sequencing (WGS) of 146 USA300 MRSA isolates from SSTIs and colonization cultures obtained from an investigation conducted from 2008 to 2010 in Chicago and Los Angeles households that included an index case with an S. aureus SSTI. Identifying unique single nucleotide polymorphisms (SNPs) and analyzing whole-genome phylogeny, we characterized isolates to understand transmission dynamics, genetic relatedness, and microevolution of USA300 MRSA within the households. We also compared the 146 USA300 MRSA isolates from our study with the previously published genome sequences of the USA300 MRSA isolates from San Diego (n = 35) and New York City (n = 277). We found little genetic variation within the USA300 MRSA household isolates from Los Angeles (mean number of SNPs ± standard deviation, 17.6 ± 35; π nucleotide diversity, 3.1 × 10(-5)) or from Chicago (mean number of SNPs ± standard deviation, 12 ± 19; π nucleotide diversity, 3.1 × 10(-5)). The isolates within a household clustered into closely related monophyletic groups, suggesting the introduction into and transmission within each household of a single common USA300 ancestral strain. From a Bayesian evolutionary reconstruction, we inferred that USA300 persisted within households for 2.33 to 8.35 years prior to sampling. We also noted that fluoroquinolone-resistant USA300 clones emerged around 1995 and were more widespread in Los Angeles and New York City than in Chicago. Our findings strongly suggest that unique USA300 MRSA isolates are transmitted within households that contain an individual with an SSTI. Decolonization of household members may be a critical component of prevention programs to control USA300 MRSA spread in the United States.
Importance: USA300, a virulent and easily transmissible strain of methicillin-resistant Staphylococcus aureus (MRSA), is the predominant community-associated MRSA clone in the United States. It most commonly causes skin infections but also causes necrotizing pneumonia and endocarditis. Strategies to limit the spread of MRSA in the community can only be effective if we understand the most common sources of transmission and the microevolutionary processes that provide a fitness advantage to MRSA. We performed a whole-genome sequence comparison of 146 USA300 MRSA isolates from Chicago and Los Angeles. We show that households represent a frequent site of transmission and a long-term reservoir of USA300 strains; individuals within households transmit the same USA300 strain among themselves. Our study also reveals that a large proportion of the USA300 isolates sequenced are resistant to fluoroquinolone antibiotics. The significance of this study is that if households serve as long-term reservoirs of USA300, household MRSA eradication programs may result in a uniquely effective control method.
Copyright © 2015 Alam et al.
Figures


Similar articles
-
Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community.Proc Natl Acad Sci U S A. 2014 May 6;111(18):6738-43. doi: 10.1073/pnas.1401006111. Epub 2014 Apr 21. Proc Natl Acad Sci U S A. 2014. PMID: 24753569 Free PMC article.
-
Persistent environmental contamination with USA300 methicillin-resistant Staphylococcus aureus and other pathogenic strain types in households with S. aureus skin infections.Infect Control Hosp Epidemiol. 2014 Nov;35(11):1373-82. doi: 10.1086/678414. Epub 2014 Sep 22. Infect Control Hosp Epidemiol. 2014. PMID: 25333432
-
Intrahost Evolution of Methicillin-Resistant Staphylococcus aureus USA300 Among Individuals With Reoccurring Skin and Soft-Tissue Infections.J Infect Dis. 2016 Sep 15;214(6):895-905. doi: 10.1093/infdis/jiw242. Epub 2016 Jun 10. J Infect Dis. 2016. PMID: 27288537 Free PMC article.
-
Virulence strategies of the dominant USA300 lineage of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA).FEMS Immunol Med Microbiol. 2012 Jun;65(1):5-22. doi: 10.1111/j.1574-695X.2012.00937.x. Epub 2012 Mar 5. FEMS Immunol Med Microbiol. 2012. PMID: 22309135 Free PMC article. Review.
-
Methicillin-resistant Staphylococcus aureus USA300 clone as a cause of Lemierre's syndrome.J Clin Microbiol. 2011 May;49(5):2063-6. doi: 10.1128/JCM.02507-10. Epub 2011 Mar 23. J Clin Microbiol. 2011. PMID: 21430106 Free PMC article. Review.
Cited by
-
Clinical outcomes and treatment approach for community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) infections in Israel.Eur J Clin Microbiol Infect Dis. 2017 Jan;36(1):153-162. doi: 10.1007/s10096-016-2789-3. Epub 2016 Sep 27. Eur J Clin Microbiol Infect Dis. 2017. PMID: 27677279
-
Antimicrobial resistance in methicillin-resistant staphylococcus aureus.Saudi J Biol Sci. 2023 Apr;30(4):103604. doi: 10.1016/j.sjbs.2023.103604. Epub 2023 Feb 28. Saudi J Biol Sci. 2023. PMID: 36936699 Free PMC article. Review.
-
Building a genomic framework for prospective MRSA surveillance in the United Kingdom and the Republic of Ireland.Genome Res. 2016 Feb;26(2):263-70. doi: 10.1101/gr.196709.115. Epub 2015 Dec 15. Genome Res. 2016. PMID: 26672018 Free PMC article.
-
Identifying the effect of patient sharing on between-hospital genetic differentiation of methicillin-resistant Staphylococcus aureus.Genome Med. 2016 Feb 13;8(1):18. doi: 10.1186/s13073-016-0274-3. Genome Med. 2016. PMID: 26873713 Free PMC article.
-
Whole genome sequencing to investigate a putative outbreak of the virulent community-associated methicillin-resistant Staphylococcus aureus ST93 clone in a remote Indigenous community.Microb Genom. 2016 Dec 12;2(12):e000098. doi: 10.1099/mgen.0.000098. eCollection 2016 Dec. Microb Genom. 2016. PMID: 28348837 Free PMC article.
References
-
- Jevons MP. 1961. Celbenin-resistant staphylococci. BMJ 1:124–125.
-
- Talan DA, Krishnadasan A, Gorwitz RJ, Fosheim GE, Limbago B, Albrecht V, Moran GJ, EMERGEncy ID Net Study Group . 2011. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin Infect Dis 53:144–149. doi:10.1093/cid/cir308. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

