Wash interacts with lamin and affects global nuclear organization
- PMID: 25754639
- PMCID: PMC4366290
- DOI: 10.1016/j.cub.2015.01.052
Wash interacts with lamin and affects global nuclear organization
Abstract
The cytoplasmic functions of Wiskott-Aldrich syndrome family (WAS) proteins are well established and include roles in cytoskeleton reorganization and membrane-cytoskeletal interactions important for membrane/vesicle trafficking, morphogenesis, immune response, and signal transduction. Misregulation of these proteins is associated with immune deficiency and metastasis [1-4]. Cytoplasmic WAS proteins act as effectors of Rho family GTPases and polymerize branched actin through the Arp2/3 complex [1, 5]. Previously, we identified Drosophila washout (wash) as a new member of the WAS family with essential cytoplasmic roles in early development [6, 7]. Studies in mammalian cells and Dictyostelium suggest that WASH functions primarily in a multiprotein complex that regulates endosome shape and trafficking in an Arp2/3-dependent manner [8-11]. However, roles for classically cytoplasmic proteins in the nucleus are beginning to emerge, in particular, as participants in the regulation of gene expression [12, 13]. Here, we show that Drosophila Wash is present in the nucleus, where it plays a key role in global nuclear organization. wash mutant and knockdown nuclei disrupt subnuclear structures/organelles and exhibit the abnormal wrinkled morphology reminiscent of those observed in diverse laminopathies [14-16]. We find that nuclear Wash interacts with B-type Lamin (Lamin Dm0), and, like Lamin, Wash associates with constitutive heterochromatin. Wash knockdown increases chromatin accessibility of repressive compartments and results in a global redistribution of repressive histone modifications. Thus, our results reveal a novel role for Wash in modulating nucleus morphology and in the organization of both chromatin and non-chromatin nuclear sub-structures.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
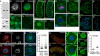
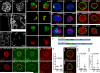

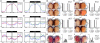
Comment in
-
Wiskott-Aldrich syndrome proteins in the nucleus: aWASH with possibilities.Nucleus. 2015;6(5):349-59. doi: 10.1080/19491034.2015.1086051. Nucleus. 2015. PMID: 26305109 Free PMC article.
Similar articles
-
Wiskott-Aldrich syndrome proteins in the nucleus: aWASH with possibilities.Nucleus. 2015;6(5):349-59. doi: 10.1080/19491034.2015.1086051. Nucleus. 2015. PMID: 26305109 Free PMC article.
-
The JIL-1 kinase interacts with lamin Dm0 and regulates nuclear lamina morphology of Drosophila nurse cells.J Cell Sci. 2005 Nov 1;118(Pt 21):5079-87. doi: 10.1242/jcs.02611. J Cell Sci. 2005. PMID: 16254246
-
Wash functions downstream of Rho and links linear and branched actin nucleation factors.Development. 2009 Aug;136(16):2849-60. doi: 10.1242/dev.035246. Development. 2009. PMID: 19633175 Free PMC article.
-
Role of nuclear lamins in nuclear organization, cellular signaling, and inherited diseases.Int Rev Cell Mol Biol. 2008;266:157-206. doi: 10.1016/S1937-6448(07)66004-3. Int Rev Cell Mol Biol. 2008. PMID: 18544494 Review.
-
Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation.Annu Rev Biochem. 2015;84:131-64. doi: 10.1146/annurev-biochem-060614-034115. Epub 2015 Feb 26. Annu Rev Biochem. 2015. PMID: 25747401 Review.
Cited by
-
Nuclear envelope budding: Getting large macromolecular complexes out of the nucleus.Bioessays. 2024 Feb;46(2):e2300182. doi: 10.1002/bies.202300182. Epub 2023 Dec 3. Bioessays. 2024. PMID: 38044581 Free PMC article.
-
Branching out in different directions: Emerging cellular functions for the Arp2/3 complex and WASP-family actin nucleation factors.Eur J Cell Biol. 2023 Jun;102(2):151301. doi: 10.1016/j.ejcb.2023.151301. Epub 2023 Mar 2. Eur J Cell Biol. 2023. PMID: 36907023 Free PMC article.
-
Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins.Mol Biol Cell. 2018 Jan 15;29(2):220-233. doi: 10.1091/mbc.E17-06-0410. Epub 2017 Nov 15. Mol Biol Cell. 2018. PMID: 29142071 Free PMC article.
-
Cellular functions of WASP family proteins at a glance.J Cell Sci. 2017 Jul 15;130(14):2235-2241. doi: 10.1242/jcs.199570. Epub 2017 Jun 23. J Cell Sci. 2017. PMID: 28646090 Free PMC article. Review.
-
Nuclear peripheral chromatin-lamin B1 interaction is required for global integrity of chromatin architecture and dynamics in human cells.Protein Cell. 2022 Apr;13(4):258-280. doi: 10.1007/s13238-020-00794-8. Epub 2020 Nov 5. Protein Cell. 2022. PMID: 33155082 Free PMC article.
References
-
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nature reviews. Molecular cell biology. 2007;8:37–48. - PubMed
-
- Rottner K, Hanisch J, Campellone KG. WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends in cell biology. 2010;20:650–661. - PubMed
-
- Massaad MJ, Ramesh N, Geha RS. Wiskott-Aldrich syndrome: a comprehensive review. Annals of the New York Academy of Sciences. 2013;1285:26–43. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

