Chromatin organization at the nuclear pore favours HIV replication
- PMID: 25744187
- PMCID: PMC4366494
- DOI: 10.1038/ncomms7483
Chromatin organization at the nuclear pore favours HIV replication
Abstract
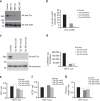
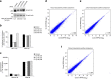
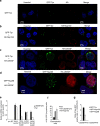
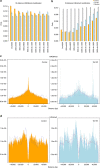
The molecular mechanisms that allow HIV to integrate into particular sites of the host genome are poorly understood. Here we tested if the nuclear pore complex (NPC) facilitates the targeting of HIV integration by acting on chromatin topology. We show that the integrity of the nuclear side of the NPC, which is mainly composed of Tpr, is not required for HIV nuclear import, but that Nup153 is essential. Depletion of Tpr markedly reduces HIV infectivity, but not the level of integration. HIV integration sites in Tpr-depleted cells are less associated with marks of active genes, consistent with the state of chromatin proximal to the NPC, as analysed by super-resolution microscopy. LEDGF/p75, which promotes viral integration into active genes, stabilizes Tpr at the nuclear periphery and vice versa. Our data support a model in which HIV nuclear import and integration are concerted steps, and where Tpr maintains a chromatin environment favourable for HIV replication.
Figures

Similar articles
-
Nuclear architecture dictates HIV-1 integration site selection.Nature. 2015 May 14;521(7551):227-31. doi: 10.1038/nature14226. Epub 2015 Mar 2. Nature. 2015. PMID: 25731161
-
A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin.Proc Natl Acad Sci U S A. 2016 Feb 23;113(8):E1054-63. doi: 10.1073/pnas.1524213113. Epub 2016 Feb 8. Proc Natl Acad Sci U S A. 2016. PMID: 26858452 Free PMC article.
-
The Structure-Specific Recognition Protein 1 Associates with Lens Epithelium-Derived Growth Factor Proteins and Modulates HIV-1 Replication.J Mol Biol. 2016 Jul 17;428(14):2814-31. doi: 10.1016/j.jmb.2016.05.013. Epub 2016 May 21. J Mol Biol. 2016. PMID: 27216501 Free PMC article.
-
Factors that mold the nuclear landscape of HIV-1 integration.Nucleic Acids Res. 2021 Jan 25;49(2):621-635. doi: 10.1093/nar/gkaa1207. Nucleic Acids Res. 2021. PMID: 33337475 Free PMC article. Review.
-
New insights in the role of nucleoporins: a bridge leading to concerted steps from HIV-1 nuclear entry until integration.Virus Res. 2013 Dec 26;178(2):187-96. doi: 10.1016/j.virusres.2013.09.003. Epub 2013 Sep 16. Virus Res. 2013. PMID: 24051001 Review.
Cited by
-
DNA minicircles clarify the specific role of DNA structure on retroviral integration.Nucleic Acids Res. 2016 Sep 19;44(16):7830-47. doi: 10.1093/nar/gkw651. Epub 2016 Jul 20. Nucleic Acids Res. 2016. PMID: 27439712 Free PMC article.
-
Strategies for Targeting Retroviral Integration for Safer Gene Therapy: Advances and Challenges.Front Mol Biosci. 2021 May 12;8:662331. doi: 10.3389/fmolb.2021.662331. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055882 Free PMC article. Review.
-
Role of Transportin-SR2 in HIV-1 Nuclear Import.Viruses. 2021 May 4;13(5):829. doi: 10.3390/v13050829. Viruses. 2021. PMID: 34064404 Free PMC article. Review.
-
Nuclear pore heterogeneity influences HIV-1 infection and the antiviral activity of MX2.Elife. 2018 Aug 7;7:e35738. doi: 10.7554/eLife.35738. Elife. 2018. PMID: 30084827 Free PMC article.
-
Defective HIV-1 genomes and their potential impact on HIV pathogenesis.Retrovirology. 2022 Jun 28;19(1):13. doi: 10.1186/s12977-022-00601-8. Retrovirology. 2022. PMID: 35764966 Free PMC article. Review.
References
-
- Kalverda B., Pickersgill H., Shloma V. V. & Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140, 360–371 (2010) . - PubMed
-
- Mendjan S. et al.. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 21, 811–823 (2006) . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

