Quantitative evolutionary dynamics using high-resolution lineage tracking
- PMID: 25731169
- PMCID: PMC4426284
- DOI: 10.1038/nature14279
Quantitative evolutionary dynamics using high-resolution lineage tracking
Abstract
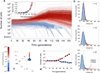
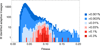
Evolution of large asexual cell populations underlies ∼30% of deaths worldwide, including those caused by bacteria, fungi, parasites, and cancer. However, the dynamics underlying these evolutionary processes remain poorly understood because they involve many competing beneficial lineages, most of which never rise above extremely low frequencies in the population. To observe these normally hidden evolutionary dynamics, we constructed a sequencing-based ultra high-resolution lineage tracking system in Saccharomyces cerevisiae that allowed us to monitor the relative frequencies of ∼500,000 lineages simultaneously. In contrast to some expectations, we found that the spectrum of fitness effects of beneficial mutations is neither exponential nor monotonic. Early adaptation is a predictable consequence of this spectrum and is strikingly reproducible, but the initial small-effect mutations are soon outcompeted by rarer large-effect mutations that result in variability between replicates. These results suggest that early evolutionary dynamics may be deterministic for a period of time before stochastic effects become important.
Figures





Comment in
-
Evolution: Fitness tracking for adapting populations.Nature. 2015 Mar 12;519(7542):164-5. doi: 10.1038/nature14207. Epub 2015 Feb 25. Nature. 2015. PMID: 25731163 No abstract available.
Similar articles
-
Beyond genome sequencing: lineage tracking with barcodes to study the dynamics of evolution, infection, and cancer.Genomics. 2014 Dec;104(6 Pt A):417-30. doi: 10.1016/j.ygeno.2014.09.005. Epub 2014 Sep 28. Genomics. 2014. PMID: 25260907 Review.
-
High-resolution lineage tracking reveals travelling wave of adaptation in laboratory yeast.Nature. 2019 Nov;575(7783):494-499. doi: 10.1038/s41586-019-1749-3. Epub 2019 Nov 13. Nature. 2019. PMID: 31723263 Free PMC article.
-
Sex speeds adaptation by altering the dynamics of molecular evolution.Nature. 2016 Mar 10;531(7593):233-6. doi: 10.1038/nature17143. Epub 2016 Feb 24. Nature. 2016. PMID: 26909573 Free PMC article.
-
Evolution: Fitness tracking for adapting populations.Nature. 2015 Mar 12;519(7542):164-5. doi: 10.1038/nature14207. Epub 2015 Feb 25. Nature. 2015. PMID: 25731163 No abstract available.
-
Cellular barcoding: lineage tracing, screening and beyond.Nat Methods. 2018 Nov;15(11):871-879. doi: 10.1038/s41592-018-0185-x. Epub 2018 Oct 30. Nat Methods. 2018. PMID: 30377352 Review.
Cited by
-
High-throughput evaluation of synthetic metabolic pathways.Technology (Singap World Sci). 2016 Mar;4(1):9-14. doi: 10.1142/S233954781640001X. Epub 2015 Dec 16. Technology (Singap World Sci). 2016. PMID: 27453919 Free PMC article.
-
Rediversification following ecotype isolation reveals hidden adaptive potential.Curr Biol. 2024 Feb 26;34(4):855-867.e6. doi: 10.1016/j.cub.2024.01.029. Epub 2024 Feb 6. Curr Biol. 2024. PMID: 38325377
-
Divergent Evolution of Mutation Rates and Biases in the Long-Term Evolution Experiment with Escherichia coli.Genome Biol Evol. 2020 Sep 1;12(9):1591-1603. doi: 10.1093/gbe/evaa178. Genome Biol Evol. 2020. PMID: 32853353 Free PMC article.
-
Tempo and mode of genome evolution in a 50,000-generation experiment.Nature. 2016 Aug 11;536(7615):165-70. doi: 10.1038/nature18959. Epub 2016 Aug 1. Nature. 2016. PMID: 27479321 Free PMC article.
-
Precise measurement of molecular phenotypes with barcode-based CRISPRi systems.bioRxiv [Preprint]. 2024 Jun 22:2024.06.21.600132. doi: 10.1101/2024.06.21.600132. bioRxiv. 2024. PMID: 38948701 Free PMC article. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

