Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation
- PMID: 25713363
- PMCID: PMC4364211
- DOI: 10.1073/pnas.1501199112
Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation
Abstract
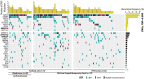
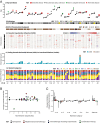
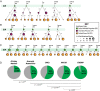
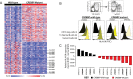
Follicular lymphoma (FL) is incurable with conventional therapies and has a clinical course typified by multiple relapses after therapy. These tumors are genetically characterized by B-cell leukemia/lymphoma 2 (BCL2) translocation and mutation of genes involved in chromatin modification. By analyzing purified tumor cells, we identified additional novel recurrently mutated genes and confirmed mutations of one or more chromatin modifier genes within 96% of FL tumors and two or more in 76% of tumors. We defined the hierarchy of somatic mutations arising during tumor evolution by analyzing the phylogenetic relationship of somatic mutations across the coding genomes of 59 sequentially acquired biopsies from 22 patients. Among all somatically mutated genes, CREBBP mutations were most significantly enriched within the earliest inferable progenitor. These mutations were associated with a signature of decreased antigen presentation characterized by reduced transcript and protein abundance of MHC class II on tumor B cells, in line with the role of CREBBP in promoting class II transactivator (CIITA)-dependent transcriptional activation of these genes. CREBBP mutant B cells stimulated less proliferation of T cells in vitro compared with wild-type B cells from the same tumor. Transcriptional signatures of tumor-infiltrating T cells were indicative of reduced proliferation, and this corresponded to decreased frequencies of tumor-infiltrating CD4 helper T cells and CD8 memory cytotoxic T cells. These observations therefore implicate CREBBP mutation as an early event in FL evolution that contributes to immune evasion via decreased antigen presentation.
Keywords: CREBBP; antigen presentation; exome; hierarchy; lymphoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The CREBBP Acetyltransferase Is a Haploinsufficient Tumor Suppressor in B-cell Lymphoma.Cancer Discov. 2017 Mar;7(3):322-337. doi: 10.1158/2159-8290.CD-16-1417. Epub 2017 Jan 9. Cancer Discov. 2017. PMID: 28069569 Free PMC article.
-
Pan-HDAC Inhibitors Restore PRDM1 Response to IL21 in CREBBP-Mutated Follicular Lymphoma.Clin Cancer Res. 2019 Jan 15;25(2):735-746. doi: 10.1158/1078-0432.CCR-18-1153. Epub 2018 Oct 22. Clin Cancer Res. 2019. PMID: 30348636
-
Crebbp loss cooperates with Bcl2 overexpression to promote lymphoma in mice.Blood. 2017 May 11;129(19):2645-2656. doi: 10.1182/blood-2016-08-733469. Epub 2017 Mar 13. Blood. 2017. PMID: 28288979 Free PMC article.
-
Pathogenesis of follicular lymphoma.Best Pract Res Clin Haematol. 2018 Mar;31(1):2-14. doi: 10.1016/j.beha.2017.10.006. Epub 2017 Nov 1. Best Pract Res Clin Haematol. 2018. PMID: 29452662 Review.
-
Chromatin modifying gene mutations in follicular lymphoma.Blood. 2018 Feb 8;131(6):595-604. doi: 10.1182/blood-2017-08-737361. Epub 2017 Nov 20. Blood. 2018. PMID: 29158360 Free PMC article. Review.
Cited by
-
DNA Methylation, Deamination, and Translesion Synthesis Combine to Generate Footprint Mutations in Cancer Driver Genes in B-Cell Derived Lymphomas and Other Cancers.Front Genet. 2021 May 19;12:671866. doi: 10.3389/fgene.2021.671866. eCollection 2021. Front Genet. 2021. PMID: 34093666 Free PMC article.
-
A human lymphoma organoid model for evaluating and targeting the follicular lymphoma tumor immune microenvironment.Cell Stem Cell. 2024 Mar 7;31(3):410-420.e4. doi: 10.1016/j.stem.2024.01.012. Epub 2024 Feb 22. Cell Stem Cell. 2024. PMID: 38402619
-
Selective Inhibition of HDAC3 Targets Synthetic Vulnerabilities and Activates Immune Surveillance in Lymphoma.Cancer Discov. 2020 Mar;10(3):440-459. doi: 10.1158/2159-8290.CD-19-0116. Epub 2020 Jan 8. Cancer Discov. 2020. PMID: 31915197 Free PMC article.
-
SCADIE: simultaneous estimation of cell type proportions and cell type-specific gene expressions using SCAD-based iterative estimating procedure.Genome Biol. 2022 Jun 15;23(1):129. doi: 10.1186/s13059-022-02688-w. Genome Biol. 2022. PMID: 35706040 Free PMC article.
-
Follicular lymphoma: updates for pathologists.J Pathol Transl Med. 2022 Jan;56(1):1-15. doi: 10.4132/jptm.2021.09.29. Epub 2021 Dec 27. J Pathol Transl Med. 2022. PMID: 34942689 Free PMC article. Review.
References
-
- Dave SS, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351(21):2159–2169. - PubMed
-
- Glas AM, et al. Gene-expression and immunohistochemical study of specific T-cell subsets and accessory cell types in the transformation and prognosis of follicular lymphoma. J Clin Oncol. 2007;25(4):390–398. - PubMed
-
- Green MR, Alizadeh AA. Common progenitor cells in mature B-cell malignancies: Implications for therapy. Curr Opin Hematol. 2014;21(4):333–340. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

