The RNA-binding protein HuR is essential for the B cell antibody response
- PMID: 25706746
- PMCID: PMC4479220
- DOI: 10.1038/ni.3115
The RNA-binding protein HuR is essential for the B cell antibody response
Abstract
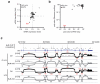
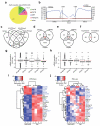
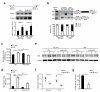
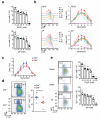
Post-transcriptional regulation of mRNA by the RNA-binding protein HuR (encoded by Elavl1) is required in B cells for the germinal center reaction and for the production of class-switched antibodies in response to thymus-independent antigens. Transcriptome-wide examination of RNA isoforms and their abundance and translation in HuR-deficient B cells, together with direct measurements of HuR-RNA interactions, revealed that HuR-dependent splicing of mRNA affected hundreds of transcripts, including that encoding dihydrolipoamide S-succinyltransferase (Dlst), a subunit of the 2-oxoglutarate dehydrogenase (α-KGDH) complex. In the absence of HuR, defective mitochondrial metabolism resulted in large amounts of reactive oxygen species and B cell death. Our study shows how post-transcriptional processes control the balance of energy metabolism required for the proliferation and differentiation of B cells.
Figures



Similar articles
-
B Cell-Intrinsic Expression of the HuR RNA-Binding Protein Is Required for the T Cell-Dependent Immune Response In Vivo.J Immunol. 2015 Oct 1;195(7):3449-62. doi: 10.4049/jimmunol.1500512. Epub 2015 Aug 28. J Immunol. 2015. PMID: 26320247 Free PMC article.
-
Swiprosin-1/EFhd2 limits germinal center responses and humoral type 2 immunity.Eur J Immunol. 2014 Nov;44(11):3206-19. doi: 10.1002/eji.201444479. Epub 2014 Sep 16. Eur J Immunol. 2014. PMID: 25092375
-
Essential role of immobilized chemokine CXCL12 in the regulation of the humoral immune response.Proc Natl Acad Sci U S A. 2017 Feb 28;114(9):2319-2324. doi: 10.1073/pnas.1611958114. Epub 2017 Feb 13. Proc Natl Acad Sci U S A. 2017. PMID: 28193885 Free PMC article.
-
The primary germinal center response in mice.Curr Opin Immunol. 2005 Jun;17(3):298-302. doi: 10.1016/j.coi.2005.04.007. Curr Opin Immunol. 2005. PMID: 15886120 Review.
-
Regulation of the mRNA-binding protein HuR by posttranslational modification: spotlight on phosphorylation.Curr Protein Pept Sci. 2012 Jun;13(4):380-90. doi: 10.2174/138920312801619439. Curr Protein Pept Sci. 2012. PMID: 22708484 Review.
Cited by
-
RNA-Binding Protein HuR Promotes Th17 Cell Differentiation and Can Be Targeted to Reduce Autoimmune Neuroinflammation.J Immunol. 2020 Apr 15;204(8):2076-2087. doi: 10.4049/jimmunol.1900769. Epub 2020 Mar 13. J Immunol. 2020. PMID: 32169842 Free PMC article.
-
Redox regulation of immunometabolism.Nat Rev Immunol. 2021 Jun;21(6):363-381. doi: 10.1038/s41577-020-00478-8. Epub 2020 Dec 18. Nat Rev Immunol. 2021. PMID: 33340021 Review.
-
Understanding and targeting the disease-related RNA binding protein human antigen R (HuR).Wiley Interdiscip Rev RNA. 2020 May;11(3):e1581. doi: 10.1002/wrna.1581. Epub 2020 Jan 23. Wiley Interdiscip Rev RNA. 2020. PMID: 31970930 Free PMC article. Review.
-
Genome-wide assessment of differential translations with ribosome profiling data.Nat Commun. 2016 Apr 4;7:11194. doi: 10.1038/ncomms11194. Nat Commun. 2016. PMID: 27041671 Free PMC article.
-
HuR controls apoptosis and activation response without effects on cytokine 3' UTRs.RNA Biol. 2019 May;16(5):686-695. doi: 10.1080/15476286.2019.1582954. Epub 2019 Mar 4. RNA Biol. 2019. PMID: 30777501 Free PMC article.
References
-
- Blair D, Dufort FJ, Chiles TC. Protein kinase Cbeta is critical for the metabolic switch to glycolysis following B-cell antigen receptor engagement. The Biochemical journal. 2012;448:165–169. - PubMed
-
- Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Seminars in immunology. 2011;23:341–349. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
- BBS/E/B/000C0409/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J001457/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L009986/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P01 AG014930/AG/NIA NIH HHS/United States
- BBS/E/B/000C0407/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

