Transfection of Sclerotinia sclerotiorum with in vitro transcripts of a naturally occurring interspecific recombinant of Sclerotinia sclerotiorum hypovirus 2 significantly reduces virulence of the fungus
- PMID: 25694604
- PMCID: PMC4403457
- DOI: 10.1128/JVI.03199-14
Transfection of Sclerotinia sclerotiorum with in vitro transcripts of a naturally occurring interspecific recombinant of Sclerotinia sclerotiorum hypovirus 2 significantly reduces virulence of the fungus
Abstract
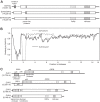
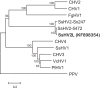
A recombinant strain of Sclerotinia sclerotiorum hypovirus 2 (SsHV2) was identified from a North American Sclerotinia sclerotiorum isolate (328) from lettuce (Lactuca sativa L.) by high-throughput sequencing of total RNA. The 5'- and 3'-terminal regions of the genome were determined by rapid amplification of cDNA ends. The assembled nucleotide sequence was up to 92% identical to two recently reported SsHV2 strains but contained a deletion near its 5' terminus of more than 1.2 kb relative to the other SsHV2 strains and an insertion of 524 nucleotides (nt) that was distantly related to Valsa ceratosperma hypovirus 1. This suggests that the new isolate is a heterologous recombinant of SsHV2 with a yet-uncharacterized hypovirus. We named the new strain Sclerotinia sclerotiorum hypovirus 2 Lactuca (SsHV2L) and deposited the sequence in GenBank with accession number KF898354. Sclerotinia sclerotiorum isolate 328 was coinfected with a strain of Sclerotinia sclerotiorum endornavirus 1 and was debilitated compared to cultures of the same isolate that had been cured of virus infection by cycloheximide treatment and hyphal tipping. To determine whether SsHV2L alone could induce hypovirulence in S. sclerotiorum, a full-length cDNA of the 14,538-nt viral genome was cloned. Transcripts corresponding to the viral RNA were synthesized in vitro and transfected into a virus-free isolate of S. sclerotiorum, DK3. Isolate DK3 transfected with SsHV2L was hypovirulent on soybean and lettuce and exhibited delayed maturation of sclerotia relative to virus-free DK3, completing Koch's postulates for the association of hypovirulence with SsHV2L.
Importance: A cosmopolitan fungus, Sclerotinia sclerotiorum infects more than 400 plant species and causes a plant disease known as white mold that produces significant yield losses in major crops annually. Mycoviruses have been used successfully to reduce losses caused by fungal plant pathogens, but definitive relationships between hypovirus infections and hypovirulence in S. sclerotiorum were lacking. By establishing a cause-and-effect relationship between Sclerotinia sclerotiorum hypovirus Lactuca (SsHV2L) infection and the reduction in host virulence, we showed direct evidence that hypoviruses have the potential to reduce the severity of white mold disease. In addition to intraspecific recombination, this study showed that recent interspecific recombination is an important factor shaping viral genomes. The construction of an infectious clone of SsHV2L allows future exploration of the interactions between SsHV2L and S. sclerotiorum, a widespread fungal pathogen of plants.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Transcriptional and Small RNA Responses of the White Mold Fungus Sclerotinia sclerotiorum to Infection by a Virulence-Attenuating Hypovirus.Viruses. 2018 Dec 14;10(12):713. doi: 10.3390/v10120713. Viruses. 2018. PMID: 30558121 Free PMC article.
-
Characterisation of a novel hypovirus from Sclerotinia sclerotiorum potentially representing a new genus within the Hypoviridae.Virology. 2014 Sep;464-465:441-449. doi: 10.1016/j.virol.2014.07.005. Epub 2014 Aug 6. Virology. 2014. PMID: 25108682
-
Co-infection of a hypovirulent isolate of Sclerotinia sclerotiorum with a new botybirnavirus and a strain of a mitovirus.Virol J. 2016 Jun 6;13:92. doi: 10.1186/s12985-016-0550-2. Virol J. 2016. PMID: 27267756 Free PMC article.
-
Viruses of the plant pathogenic fungus Sclerotinia sclerotiorum.Adv Virus Res. 2013;86:215-48. doi: 10.1016/B978-0-12-394315-6.00008-8. Adv Virus Res. 2013. PMID: 23498908 Review.
-
Mycovirus-induced hypovirulence in notorious fungi Sclerotinia: a comprehensive review.Braz J Microbiol. 2023 Sep;54(3):1459-1478. doi: 10.1007/s42770-023-01073-4. Epub 2023 Jul 31. Braz J Microbiol. 2023. PMID: 37523037 Free PMC article. Review.
Cited by
-
Identification of Diverse Mycoviruses through Metatranscriptomics Characterization of the Viromes of Five Major Fungal Plant Pathogens.J Virol. 2016 Jul 11;90(15):6846-6863. doi: 10.1128/JVI.00357-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27194764 Free PMC article.
-
Molecular characterization of a bipartite double-stranded RNA virus and its satellite-like RNA co-infecting the phytopathogenic fungus Sclerotinia sclerotiorum.Front Microbiol. 2015 May 6;6:406. doi: 10.3389/fmicb.2015.00406. eCollection 2015. Front Microbiol. 2015. PMID: 25999933 Free PMC article.
-
Diverse, Novel Mycoviruses From the Virome of a Hypovirulent Sclerotium rolfsii Strain.Front Plant Sci. 2018 Nov 27;9:1738. doi: 10.3389/fpls.2018.01738. eCollection 2018. Front Plant Sci. 2018. PMID: 30542362 Free PMC article.
-
Alphapartitiviruses of Heterobasidion Wood Decay Fungi Affect Each Other's Transmission and Host Growth.Front Cell Infect Microbiol. 2019 Mar 26;9:64. doi: 10.3389/fcimb.2019.00064. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 30972301 Free PMC article.
-
Establishment of Neurospora crassa as a model organism for fungal virology.Nat Commun. 2020 Nov 6;11(1):5627. doi: 10.1038/s41467-020-19355-y. Nat Commun. 2020. PMID: 33159072 Free PMC article.
References
-
- Boland GJ. 2004. Fungal viruses, hypovirulence, and biological control of Sclerotinia species. Can J Plant Pathol 26:6–18. doi:10.1080/07060660409507107. - DOI
-
- Purdy LH. 1979. Sclerotinia sclerotiorum: history, diseases and symptomatology, host range, geographic distribution, and impact. Phytopathology 69:875–880. doi:10.1094/Phyto-69-875. - DOI
-
- Steadman JR. 1979. Control of plant-diseases caused by Sclerotinia species. Phytopathology 69:904–907. doi:10.1094/Phyto-69-904. - DOI
-
- Bardin SD, Huang HC. 2001. Research on biology and control of Sclerotinia diseases in Canada. Can J Plant Pathol 23:88–98. doi:10.1080/07060660109506914. - DOI
-
- Boland GJ, Hall R. 1994. Index of plant hosts of Sclerotinia sclerotiorum. Can J Plant Pathol 16:93–108. doi:10.1080/07060669409500766. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

