Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation
- PMID: 25686250
- PMCID: PMC4344897
- DOI: 10.1038/ncb3109
Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation
Erratum in
-
Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation.Nat Cell Biol. 2015 Apr;17(4):531. doi: 10.1038/ncb3155. Nat Cell Biol. 2015. PMID: 25812525 No abstract available.
Abstract
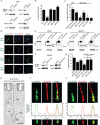
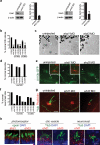
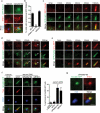
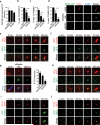
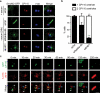
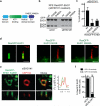
Membrane association with mother centriole (M-centriole) distal appendages is critical for ciliogenesis initiation. How the Rab GTPase Rab11-Rab8 cascade functions in early ciliary membrane assembly is unknown. Here, we show that the membrane shaping proteins EHD1 and EHD3, in association with the Rab11-Rab8 cascade, function in early ciliogenesis. EHD1 and EHD3 localize to preciliary membranes and the ciliary pocket. EHD-dependent membrane tubulation is essential for ciliary vesicle formation from smaller distal appendage vesicles (DAVs). Importantly, this step functions in M-centriole to basal body transformation and recruitment of transition zone proteins and IFT20. SNAP29, a SNARE membrane fusion regulator and EHD1-binding protein, is also required for DAV-mediated ciliary vesicle assembly. Interestingly, only after ciliary vesicle assembly is Rab8 activated for ciliary growth. Our studies uncover molecular mechanisms informing a previously uncharacterized ciliogenesis step, whereby EHD1 and EHD3 reorganize the M-centriole and associated DAVs before coordinated ciliary membrane and axoneme growth.
Figures


Comment in
-
Ciliary vesicle formation: a prelude to ciliogenesis.Dev Cell. 2015 Mar 23;32(6):665-6. doi: 10.1016/j.devcel.2015.03.012. Dev Cell. 2015. PMID: 25805133 Free PMC article.
Similar articles
-
The novel centriolar satellite protein SSX2IP targets Cep290 to the ciliary transition zone.Mol Biol Cell. 2014 Feb;25(4):495-507. doi: 10.1091/mbc.E13-09-0526. Epub 2013 Dec 19. Mol Biol Cell. 2014. PMID: 24356449 Free PMC article.
-
Myosin-Va is required for preciliary vesicle transportation to the mother centriole during ciliogenesis.Nat Cell Biol. 2018 Feb;20(2):175-185. doi: 10.1038/s41556-017-0018-7. Epub 2018 Jan 15. Nat Cell Biol. 2018. PMID: 29335527
-
Investigation of F-BAR domain PACSIN proteins uncovers membrane tubulation function in cilia assembly and transport.Nat Commun. 2019 Jan 25;10(1):428. doi: 10.1038/s41467-018-08192-9. Nat Commun. 2019. PMID: 30683896 Free PMC article.
-
[The ciliary pocket: a rendezvous between the centrosome and vesicular trafficking].Med Sci (Paris). 2014 Nov;30(11):962-7. doi: 10.1051/medsci/20143011009. Epub 2014 Nov 10. Med Sci (Paris). 2014. PMID: 25388577 Review. French.
-
Ciliogenesis membrane dynamics and organization.Semin Cell Dev Biol. 2023 Jan 15;133:20-31. doi: 10.1016/j.semcdb.2022.03.021. Epub 2022 Mar 26. Semin Cell Dev Biol. 2023. PMID: 35351373 Free PMC article. Review.
Cited by
-
IFT20: An Eclectic Regulator of Cellular Processes beyond Intraflagellar Transport.Int J Mol Sci. 2022 Oct 12;23(20):12147. doi: 10.3390/ijms232012147. Int J Mol Sci. 2022. PMID: 36292997 Free PMC article. Review.
-
The IN/OUT assay: a new tool to study ciliogenesis.Cilia. 2016 Aug 4;5:23. doi: 10.1186/s13630-016-0044-2. eCollection 2016. Cilia. 2016. PMID: 27493724 Free PMC article.
-
A ciliopathy complex builds distal appendages to initiate ciliogenesis.J Cell Biol. 2021 Sep 6;220(9):e202011133. doi: 10.1083/jcb.202011133. Epub 2021 Jul 9. J Cell Biol. 2021. PMID: 34241634 Free PMC article.
-
Centriole splitting caused by loss of the centrosomal linker protein C-NAP1 reduces centriolar satellite density and impedes centrosome amplification.Mol Biol Cell. 2017 Mar 15;28(6):736-745. doi: 10.1091/mbc.E16-05-0325. Epub 2017 Jan 18. Mol Biol Cell. 2017. PMID: 28100636 Free PMC article.
-
CEP290 is essential for the initiation of ciliary transition zone assembly.PLoS Biol. 2020 Dec 28;18(12):e3001034. doi: 10.1371/journal.pbio.3001034. eCollection 2020 Dec. PLoS Biol. 2020. Update in: PLoS Biol. 2024 Mar 5;22(3):e3002330. doi: 10.1371/journal.pbio.3002330. PMID: 33370260 Free PMC article. Updated.
References
-
- Ishikawa H, Kubo A, Tsukita S, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 2005;7:517–524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

