Human adenovirus 52 uses sialic acid-containing glycoproteins and the coxsackie and adenovirus receptor for binding to target cells
- PMID: 25674795
- PMCID: PMC4335501
- DOI: 10.1371/journal.ppat.1004657
Human adenovirus 52 uses sialic acid-containing glycoproteins and the coxsackie and adenovirus receptor for binding to target cells
Abstract
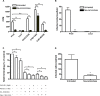
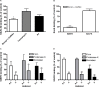
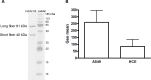
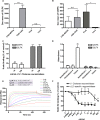
Most adenoviruses attach to host cells by means of the protruding fiber protein that binds to host cells via the coxsackievirus and adenovirus receptor (CAR) protein. Human adenovirus type 52 (HAdV-52) is one of only three gastroenteritis-causing HAdVs that are equipped with two different fiber proteins, one long and one short. Here we show, by means of virion-cell binding and infection experiments, that HAdV-52 can also attach to host cells via CAR, but most of the binding depends on sialylated glycoproteins. Glycan microarray, flow cytometry, surface plasmon resonance and ELISA analyses reveal that the terminal knob domain of the long fiber (52LFK) binds to CAR, and the knob domain of the short fiber (52SFK) binds to sialylated glycoproteins. X-ray crystallographic analysis of 52SFK in complex with 2-O-methylated sialic acid combined with functional studies of knob mutants revealed a new sialic acid binding site compared to other, known adenovirus:glycan interactions. Our findings shed light on adenovirus biology and may help to improve targeting of adenovirus-based vectors for gene therapy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Defining a Novel Role for the Coxsackievirus and Adenovirus Receptor in Human Adenovirus Serotype 5 Transduction In Vitro in the Presence of Mouse Serum.J Virol. 2017 May 26;91(12):e02487-16. doi: 10.1128/JVI.02487-16. Print 2017 Jun 15. J Virol. 2017. PMID: 28381574 Free PMC article.
-
Tropism and transduction of oncolytic adenovirus 5 vectors in cancer therapy: Focus on fiber chimerism and mosaicism, hexon and pIX.Virus Res. 2018 Sep 15;257:40-51. doi: 10.1016/j.virusres.2018.08.012. Epub 2018 Aug 17. Virus Res. 2018. PMID: 30125593 Review.
-
Polysialic acid is a cellular receptor for human adenovirus 52.Proc Natl Acad Sci U S A. 2018 May 1;115(18):E4264-E4273. doi: 10.1073/pnas.1716900115. Epub 2018 Apr 19. Proc Natl Acad Sci U S A. 2018. PMID: 29674446 Free PMC article.
-
Sialic Acid-Containing Glycans as Cellular Receptors for Ocular Human Adenoviruses: Implications for Tropism and Treatment.Viruses. 2019 Apr 27;11(5):395. doi: 10.3390/v11050395. Viruses. 2019. PMID: 31035532 Free PMC article.
-
Human adenovirus binding to host cell receptors: a structural view.Med Microbiol Immunol. 2020 Jun;209(3):325-333. doi: 10.1007/s00430-019-00645-2. Epub 2019 Nov 29. Med Microbiol Immunol. 2020. PMID: 31784892 Free PMC article. Review.
Cited by
-
Heparan Sulfate Is a Cellular Receptor for Enteric Human Adenoviruses.Viruses. 2021 Feb 14;13(2):298. doi: 10.3390/v13020298. Viruses. 2021. PMID: 33672966 Free PMC article.
-
Structure and Sialyllactose Binding of the Carboxy-Terminal Head Domain of the Fibre from a Siadenovirus, Turkey Adenovirus 3.PLoS One. 2015 Sep 29;10(9):e0139339. doi: 10.1371/journal.pone.0139339. eCollection 2015. PLoS One. 2015. PMID: 26418008 Free PMC article.
-
Tight Junctions Go Viral!Viruses. 2015 Sep 23;7(9):5145-54. doi: 10.3390/v7092865. Viruses. 2015. PMID: 26404354 Free PMC article. Review.
-
New Insights to Adenovirus-Directed Innate Immunity in Respiratory Epithelial Cells.Microorganisms. 2019 Jul 25;7(8):216. doi: 10.3390/microorganisms7080216. Microorganisms. 2019. PMID: 31349602 Free PMC article. Review.
-
Getting genetic access to natural adenovirus genomes to explore vector diversity.Virus Genes. 2017 Oct;53(5):675-683. doi: 10.1007/s11262-017-1487-2. Epub 2017 Jul 15. Virus Genes. 2017. PMID: 28711987 Review.
References
-
- Harrach B, Benkö M, Both GW, Brown M, Davison AJ, et al. (2011) Family Adenoviridae In: King AMQ, Adams MJ, Carstens EB, LE J, editors. Virus Taxonomy Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier Academic Press; pp. 125–141.
-
- Wold WSM, Horwitz MS (2007) Adenoviruses In: Knipe DM, Howley PM, editors. Fields Virology. 5 ed. Philadelphia: Lippincott Williams & Wilkins; pp. 2395–2436.
-
- Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, et al. (1997) Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275: 1320–1323. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

