Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation
- PMID: 25658430
- PMCID: PMC4450067
- DOI: 10.1371/journal.ppat.1004659
Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation
Abstract
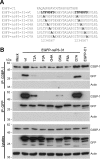
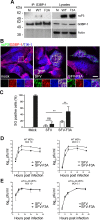
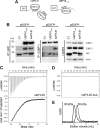
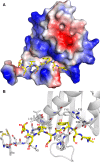
The Ras-GAP SH3 domain-binding proteins (G3BP) are essential regulators of the formation of stress granules (SG), cytosolic aggregates of proteins and RNA that are induced upon cellular stress, such as virus infection. Many viruses, including Semliki Forest virus (SFV), block SG induction by targeting G3BP. In this work, we demonstrate that the G3BP-binding motif of SFV nsP3 consists of two FGDF motifs, in which both phenylalanine and the glycine residue are essential for binding. In addition, we show that binding of the cellular G3BP-binding partner USP10 is also mediated by an FGDF motif. Overexpression of wt USP10, but not a mutant lacking the FGDF-motif, blocks SG assembly. Further, we identified FGDF-mediated G3BP binding site in herpes simplex virus (HSV) protein ICP8, and show that ICP8 binding to G3BP also inhibits SG formation, which is a novel function of HSV ICP8. We present a model of the three-dimensional structure of G3BP bound to an FGDF-containing peptide, likely representing a binding mode shared by many proteins to target G3BP.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Separate domains of G3BP promote efficient clustering of alphavirus replication complexes and recruitment of the translation initiation machinery.PLoS Pathog. 2019 Jun 14;15(6):e1007842. doi: 10.1371/journal.ppat.1007842. eCollection 2019 Jun. PLoS Pathog. 2019. PMID: 31199850 Free PMC article.
-
Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication.Open Biol. 2016 Jul;6(7):160078. doi: 10.1098/rsob.160078. Open Biol. 2016. PMID: 27383630 Free PMC article.
-
G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits.J Cell Biol. 2016 Mar 28;212(7):845-60. doi: 10.1083/jcb.201508028. J Cell Biol. 2016. PMID: 27022092 Free PMC article.
-
Research Progress on the Structure and Function of G3BP.Front Immunol. 2021 Aug 30;12:718548. doi: 10.3389/fimmu.2021.718548. eCollection 2021. Front Immunol. 2021. PMID: 34526993 Free PMC article. Review.
-
Role(s) of G3BPs in Human Pathogenesis.J Pharmacol Exp Ther. 2023 Oct;387(1):100-110. doi: 10.1124/jpet.122.001538. Epub 2023 Jul 19. J Pharmacol Exp Ther. 2023. PMID: 37468286 Free PMC article. Review.
Cited by
-
Proteome expansion in the Potyviridae evolutionary radiation.FEMS Microbiol Rev. 2022 Jul 1;46(4):fuac011. doi: 10.1093/femsre/fuac011. FEMS Microbiol Rev. 2022. PMID: 35195244 Free PMC article. Review.
-
G3BPs in Plant Stress.Front Plant Sci. 2021 Jun 10;12:680710. doi: 10.3389/fpls.2021.680710. eCollection 2021. Front Plant Sci. 2021. PMID: 34177995 Free PMC article. Review.
-
Differential Phosphatidylinositol-3-Kinase-Akt-mTOR Activation by Semliki Forest and Chikungunya Viruses Is Dependent on nsP3 and Connected to Replication Complex Internalization.J Virol. 2015 Nov;89(22):11420-37. doi: 10.1128/JVI.01579-15. Epub 2015 Sep 2. J Virol. 2015. PMID: 26339054 Free PMC article.
-
Poly(A)-binding protein is an ataxin-2 chaperone that regulates biomolecular condensates.Mol Cell. 2023 Jun 15;83(12):2020-2034.e6. doi: 10.1016/j.molcel.2023.05.025. Epub 2023 Jun 8. Mol Cell. 2023. PMID: 37295429 Free PMC article.
-
Current and Promising Antivirals Against Chikungunya Virus.Front Public Health. 2020 Dec 15;8:618624. doi: 10.3389/fpubh.2020.618624. eCollection 2020. Front Public Health. 2020. PMID: 33384981 Free PMC article. Review.
References
-
- Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, et al. (2006) Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem 281: 32870–32878. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

