Oxidative stress alters global histone modification and DNA methylation
- PMID: 25656994
- PMCID: PMC4464695
- DOI: 10.1016/j.freeradbiomed.2015.01.028
Oxidative stress alters global histone modification and DNA methylation
Abstract
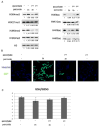
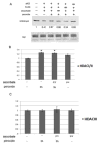
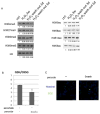
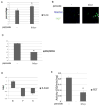
The JmjC domain-containing histone demethylases can remove histone lysine methylation and thereby regulate gene expression. The JmjC domain uses iron Fe(II) and α-ketoglutarate (αKG) as cofactors in an oxidative demethylation reaction via hydroxymethyl lysine. We hypothesize that reactive oxygen species will oxidize Fe(II) to Fe(III), thereby attenuating the activity of JmjC domain-containing histone demethylases. To minimize secondary responses from cells, extremely short periods of oxidative stress (3h) were used to investigate this question. Cells that were exposed to hydrogen peroxide (H2O2) for 3h exhibited increases in several histone methylation marks including H3K4me3 and decreases of histone acetylation marks including H3K9ac and H4K8ac; preincubation with ascorbate attenuated these changes. The oxidative stress level was measured by generation of 2',7'-dichlorofluorescein, GSH/GSSG ratio, and protein carbonyl content. A cell-free system indicated that H2O2 inhibited histone demethylase activity where increased Fe(II) rescued this inhibition. TET protein showed a decreased activity under oxidative stress. Cells exposed to a low-dose and long-term (3 weeks) oxidative stress also showed increased global levels of H3K4me3 and H3K27me3. However, these global methylation changes did not persist after washout. The cells exposed to short-term oxidative stress also appeared to have higher activity of class I/II histone deacetylase (HDAC) but not class III HDAC. In conclusion, we have found that oxidative stress transiently alters the epigenetic program process through modulating the activity of enzymes responsible for demethylation and deacetylation of histones.
Keywords: Histone demethylase; Hydrogen peroxide; Oxidative stress.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
Similar articles
-
Structural insights into histone lysine demethylation.Curr Opin Struct Biol. 2010 Dec;20(6):739-48. doi: 10.1016/j.sbi.2010.09.006. Epub 2010 Oct 21. Curr Opin Struct Biol. 2010. PMID: 20970991 Free PMC article. Review.
-
The epigenetic role of vitamin C in health and disease.Cell Mol Life Sci. 2016 Apr;73(8):1645-58. doi: 10.1007/s00018-016-2145-x. Epub 2016 Feb 4. Cell Mol Life Sci. 2016. PMID: 26846695 Free PMC article. Review.
-
Nitric oxide modifies global histone methylation by inhibiting Jumonji C domain-containing demethylases.J Biol Chem. 2013 May 31;288(22):16004-15. doi: 10.1074/jbc.M112.432294. Epub 2013 Apr 1. J Biol Chem. 2013. PMID: 23546878 Free PMC article.
-
Emerging Roles of JmjC Domain-Containing Proteins.Int Rev Cell Mol Biol. 2015;319:165-220. doi: 10.1016/bs.ircmb.2015.07.003. Epub 2015 Aug 19. Int Rev Cell Mol Biol. 2015. PMID: 26404469 Review.
-
Ascorbic Acid Reprograms Epigenome and Epitranscriptome by Reducing Fe(III) in the Catalytic Cycle of Dioxygenases.ACS Chem Biol. 2024 Jan 19;19(1):129-140. doi: 10.1021/acschembio.3c00567. Epub 2023 Dec 15. ACS Chem Biol. 2024. PMID: 38100359
Cited by
-
Methylenetetrahydrofolate reductase gene polymorphism, global DNA methylation and blood pressure: a population based study from North India.BMC Med Genomics. 2021 Feb 27;14(1):59. doi: 10.1186/s12920-021-00895-1. BMC Med Genomics. 2021. PMID: 33639933 Free PMC article.
-
Nuclear and Cytoplasmic Functions of Vitamin C.Chem Res Toxicol. 2020 Oct 19;33(10):2515-2526. doi: 10.1021/acs.chemrestox.0c00348. Epub 2020 Oct 1. Chem Res Toxicol. 2020. PMID: 33001635 Free PMC article. Review.
-
MdGGT1 Impacts Apple miR156 Precursor Levels via Ontogenetic Changes in Subcellular Glutathione Homeostasis.Front Plant Sci. 2019 Jul 31;10:994. doi: 10.3389/fpls.2019.00994. eCollection 2019. Front Plant Sci. 2019. PMID: 31417600 Free PMC article.
-
A data integration approach unveils a transcriptional signature of type 2 diabetes progression in rat and human islets.PLoS One. 2023 Oct 10;18(10):e0292579. doi: 10.1371/journal.pone.0292579. eCollection 2023. PLoS One. 2023. PMID: 37816033 Free PMC article.
-
Role of Nkx2.5 in H2O2-induced Nsd1 suppression.Cell Stress Chaperones. 2019 Jul;24(4):697-707. doi: 10.1007/s12192-019-00995-z. Epub 2019 May 18. Cell Stress Chaperones. 2019. PMID: 31104268 Free PMC article.
References
-
- Chia N, Wang L, Lu X, Senut MC, Brenner C, Ruden DM. Hypothesis: Environmental regulation of 5-hydroxymethyl-cytosine by oxidative stress. Epigenetics. 2011;6:7. - PubMed
-
- Costa M. Dioxygenases as targets of metals, hypoxia and oxidative stress during carcinogenesis. Molecular and genetic medicine. 2013;7:3.
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
-
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. - PubMed
-
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, Gaudet F, Li E, Chen T. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nature genetics. 2009;41:125–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016087/CA/NCI NIH HHS/United States
- R01 ES014454/ES/NIEHS NIH HHS/United States
- R01 ES022935/ES/NIEHS NIH HHS/United States
- R01 ES005512/ES/NIEHS NIH HHS/United States
- P30 ES000260/ES/NIEHS NIH HHS/United States
- ES023174/ES/NIEHS NIH HHS/United States
- ES010344/ES/NIEHS NIH HHS/United States
- ES014454/ES/NIEHS NIH HHS/United States
- P42 ES010344/ES/NIEHS NIH HHS/United States
- ES 022935/ES/NIEHS NIH HHS/United States
- ES005512/ES/NIEHS NIH HHS/United States
- R01 ES023174/ES/NIEHS NIH HHS/United States
- ES000260/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

