Diversification of importin-α isoforms in cellular trafficking and disease states
- PMID: 25656054
- PMCID: PMC4405237
- DOI: 10.1042/BJ20141186
Diversification of importin-α isoforms in cellular trafficking and disease states
Abstract
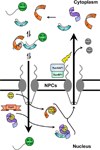
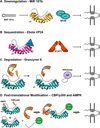
The human genome encodes seven isoforms of importin α which are grouped into three subfamilies known as α1, α2 and α3. All isoforms share a fundamentally conserved architecture that consists of an N-terminal, autoinhibitory, importin-β-binding (IBB) domain and a C-terminal Arm (Armadillo)-core that associates with nuclear localization signal (NLS) cargoes. Despite striking similarity in amino acid sequence and 3D structure, importin-α isoforms display remarkable substrate specificity in vivo. In the present review, we look at key differences among importin-α isoforms and provide a comprehensive inventory of known viral and cellular cargoes that have been shown to associate preferentially with specific isoforms. We illustrate how the diversification of the adaptor importin α into seven isoforms expands the dynamic range and regulatory control of nucleocytoplasmic transport, offering unexpected opportunities for pharmacological intervention. The emerging view of importin α is that of a key signalling molecule, with isoforms that confer preferential nuclear entry and spatiotemporal specificity on viral and cellular cargoes directly linked to human diseases.
Figures




Similar articles
-
Nuclear Respiratory Factor 2β (NRF-2β) recruits NRF-2α to the nucleus by binding to importin-α:β via an unusual monopartite-type nuclear localization signal.J Mol Biol. 2013 Sep 23;425(18):3536-48. doi: 10.1016/j.jmb.2013.07.007. Epub 2013 Jul 13. J Mol Biol. 2013. PMID: 23856623
-
An importin alpha/beta-recognized bipartite nuclear localization signal mediates targeting of the human herpes simplex virus type 1 DNA polymerase catalytic subunit pUL30 to the nucleus.Biochemistry. 2007 Aug 14;46(32):9155-63. doi: 10.1021/bi7002394. Epub 2007 Jul 20. Biochemistry. 2007. PMID: 17640102
-
Three-dimensional context rather than NLS amino acid sequence determines importin α subtype specificity for RCC1.Nat Commun. 2017 Oct 17;8(1):979. doi: 10.1038/s41467-017-01057-7. Nat Commun. 2017. PMID: 29042532 Free PMC article.
-
The importin β binding domain as a master regulator of nucleocytoplasmic transport.Biochim Biophys Acta. 2011 Sep;1813(9):1578-92. doi: 10.1016/j.bbamcr.2010.10.012. Epub 2010 Oct 26. Biochim Biophys Acta. 2011. PMID: 21029753 Free PMC article. Review.
-
Importin α: a key molecule in nuclear transport and non-transport functions.J Biochem. 2016 Aug;160(2):69-75. doi: 10.1093/jb/mvw036. Epub 2016 Jun 11. J Biochem. 2016. PMID: 27289017 Review.
Cited by
-
A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin α-dependent nuclear localization signals.Protein Sci. 2024 Feb;33(2):e4876. doi: 10.1002/pro.4876. Protein Sci. 2024. PMID: 38108201 Free PMC article.
-
Karyopherins in the Remodeling of Extracellular Matrix: Implications in Tendon Injury.J Orthop Sports Med. 2023;5(3):357-374. doi: 10.26502/josm.511500122. Epub 2023 Sep 14. J Orthop Sports Med. 2023. PMID: 37829147 Free PMC article.
-
The nuclear pore proteins Nup88/214 and T-cell acute lymphatic leukemia-associated NUP214 fusion proteins regulate Notch signaling.J Biol Chem. 2019 Aug 2;294(31):11741-11750. doi: 10.1074/jbc.RA118.006357. Epub 2019 Jun 11. J Biol Chem. 2019. PMID: 31186352 Free PMC article.
-
Terminase Large Subunit Provides a New Drug Target for Herpesvirus Treatment.Viruses. 2019 Mar 5;11(3):219. doi: 10.3390/v11030219. Viruses. 2019. PMID: 30841485 Free PMC article. Review.
-
Cell surface localization of importin α1/KPNA2 affects cancer cell proliferation by regulating FGF1 signalling.Sci Rep. 2016 Feb 18;6:21410. doi: 10.1038/srep21410. Sci Rep. 2016. PMID: 26887791 Free PMC article.
References
-
- Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. - PubMed
-
- Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. - PubMed
-
- Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays. 2000;22:532–544. - PubMed
-
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

