Expression and function of Drosophila cyclin A during embryonic cell cycle progression
- PMID: 2564316
- PMCID: PMC2753420
- DOI: 10.1016/0092-8674(89)90629-6
Expression and function of Drosophila cyclin A during embryonic cell cycle progression
Abstract
Cyclin proteins are thought to trigger entry into mitosis. During mitosis they are rapidly degraded. Therefore, mitosis and consequently cyclin degradation might be triggered at a time when cyclins have reaccumulated to a critical level. We cloned and sequenced a Drosophila cyclin A homolog and identified mutations in the corresponding gene. Immunofluorescent staining revealed that cyclin A accumulates in the interphase cytoplasm of cellularized embryos, but relocates to the nuclear region early in prophase and is completely degraded within metaphase. Cyclin A was expressed in dividing cells throughout development, and a functional cyclin A gene was required for continued division after exhaustion of maternally contributed cyclin A. Importantly, the timing of post cellularization divisions was not governed by the rate of accumulation or level of cyclin A.
Figures


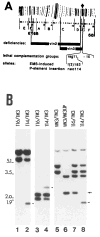
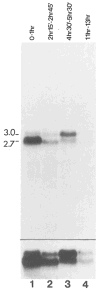

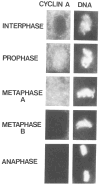
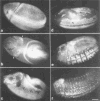
Similar articles
-
The A- and B-type cyclins of Drosophila are accumulated and destroyed in temporally distinct events that define separable phases of the G2-M transition.EMBO J. 1990 Aug;9(8):2563-72. doi: 10.1002/j.1460-2075.1990.tb07437.x. EMBO J. 1990. PMID: 2142452 Free PMC article.
-
The roles of Drosophila cyclins A and B in mitotic control.Cell. 1990 May 4;61(3):535-47. doi: 10.1016/0092-8674(90)90535-m. Cell. 1990. PMID: 2139805 Free PMC article.
-
Synergistic action of Drosophila cyclins A and B during the G2-M transition.EMBO J. 1993 Jan;12(1):65-74. doi: 10.1002/j.1460-2075.1993.tb05632.x. EMBO J. 1993. PMID: 8428595 Free PMC article.
-
Directing cell division during development.Science. 1989 Nov 3;246(4930):635-40. doi: 10.1126/science.2683080. Science. 1989. PMID: 2683080 Review.
-
Regulation of the embryonic cell proliferation by Drosophila cyclin D and cyclin E complexes.Novartis Found Symp. 2001;237:43-54; discussion 54-7, 93-9. doi: 10.1002/0470846666.ch5. Novartis Found Symp. 2001. PMID: 11444049 Review.
Cited by
-
Different cyclin types collaborate to reverse the S-phase checkpoint and permit prompt mitosis.J Cell Biol. 2012 Sep 17;198(6):973-80. doi: 10.1083/jcb.201205007. Epub 2012 Sep 10. J Cell Biol. 2012. PMID: 22965907 Free PMC article.
-
Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase.J Cell Biol. 1992 Jun;117(5):921-34. doi: 10.1083/jcb.117.5.921. J Cell Biol. 1992. PMID: 1315785 Free PMC article.
-
A screen for identifying genes interacting with armadillo, the Drosophila homolog of beta-catenin.Genetics. 1999 Dec;153(4):1753-66. doi: 10.1093/genetics/153.4.1753. Genetics. 1999. PMID: 10581282 Free PMC article.
-
Drosophila protein phosphatase V functionally complements a SIT4 mutant in Saccharomyces cerevisiae and its amino-terminal region can confer this complementation to a heterologous phosphatase catalytic domain.EMBO J. 1993 Dec;12(12):4833-42. doi: 10.1002/j.1460-2075.1993.tb06173.x. EMBO J. 1993. PMID: 8223492 Free PMC article.
-
Mutations in the Drosophila condensin subunit dCAP-G: defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase.Genetics. 2004 Oct;168(2):895-906. doi: 10.1534/genetics.104.030908. Genetics. 2004. PMID: 15514062 Free PMC article.
References
-
- Akam ME, Roberts DB, Richards GP, Ashburner M. Drosophila: the genetics of two major larval proteins. Cell. 1978;13:215–225. - PubMed
-
- Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harbor Symp Quant Biol. 1973;38:205–233. - PubMed
-
- Carroll SB, Laughon A. Production and purification of polyclonal antibodies to the foreign segment of (β-galactosidase fusion proteins. In: Glover D, editor. DNA Cloning: A Practical Approach. Oxford: IRL Press; 1987. pp. 89–111.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

