The bacterial curli system possesses a potent and selective inhibitor of amyloid formation
- PMID: 25620560
- PMCID: PMC4320674
- DOI: 10.1016/j.molcel.2014.12.025
The bacterial curli system possesses a potent and selective inhibitor of amyloid formation
Abstract
Curli are extracellular functional amyloids that are assembled by enteric bacteria during biofilm formation and host colonization. An efficient secretion system and chaperone network ensures that the major curli fiber subunit, CsgA, does not form intracellular amyloid aggregates. We discovered that the periplasmic protein CsgC was a highly effective inhibitor of CsgA amyloid formation. In the absence of CsgC, CsgA formed toxic intracellular aggregates. In vitro, CsgC inhibited CsgA amyloid formation at substoichiometric concentrations and maintained CsgA in a non-β-sheet-rich conformation. Interestingly, CsgC inhibited amyloid assembly of human α-synuclein, but not Aβ42, in vitro. We identified a common D-Q-Φ-X0,1-G-K-N-ζ-E motif in CsgC client proteins that is not found in Aβ42. CsgC is therefore both an efficient and selective amyloid inhibitor. Dedicated functional amyloid inhibitors may be a key feature that distinguishes functional amyloids from disease-associated amyloids.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
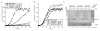
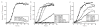
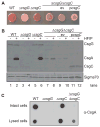



Comment in
-
Chaos controlled: discovery of a powerful amyloid inhibitor.Mol Cell. 2015 Feb 5;57(3):391-3. doi: 10.1016/j.molcel.2015.01.031. Mol Cell. 2015. PMID: 25658202
Similar articles
-
Structure-Function Analysis of the Curli Accessory Protein CsgE Defines Surfaces Essential for Coordinating Amyloid Fiber Formation.mBio. 2018 Jul 17;9(4):e01349-18. doi: 10.1128/mBio.01349-18. mBio. 2018. PMID: 30018113 Free PMC article.
-
Bacterial Chaperones CsgE and CsgC Differentially Modulate Human α-Synuclein Amyloid Formation via Transient Contacts.PLoS One. 2015 Oct 14;10(10):e0140194. doi: 10.1371/journal.pone.0140194. eCollection 2015. PLoS One. 2015. PMID: 26465894 Free PMC article.
-
Modulation of curli assembly and pellicle biofilm formation by chemical and protein chaperones.Chem Biol. 2013 Oct 24;20(10):1245-54. doi: 10.1016/j.chembiol.2013.07.017. Epub 2013 Sep 12. Chem Biol. 2013. PMID: 24035282 Free PMC article.
-
Curli provide the template for understanding controlled amyloid propagation.Prion. 2008 Apr-Jun;2(2):57-60. doi: 10.4161/pri.2.2.6746. Epub 2008 Apr 5. Prion. 2008. PMID: 19098444 Free PMC article. Review.
-
Bacterial amyloid formation: structural insights into curli biogensis.Trends Microbiol. 2015 Nov;23(11):693-706. doi: 10.1016/j.tim.2015.07.010. Epub 2015 Oct 1. Trends Microbiol. 2015. PMID: 26439293 Free PMC article. Review.
Cited by
-
Amyloid by Design: Intrinsic Regulation of Microbial Amyloid Assembly.J Mol Biol. 2018 Oct 12;430(20):3631-3641. doi: 10.1016/j.jmb.2018.07.007. Epub 2018 Jul 12. J Mol Biol. 2018. PMID: 30017921 Free PMC article. Review.
-
Assembly and substrate recognition of curli biogenesis system.Nat Commun. 2020 Jan 13;11(1):241. doi: 10.1038/s41467-019-14145-7. Nat Commun. 2020. PMID: 31932609 Free PMC article.
-
Selective inhibition of the amyloid matrix of Escherichia coli biofilms by a bifunctional microbial metabolite.NPJ Biofilms Microbiomes. 2023 Oct 19;9(1):81. doi: 10.1038/s41522-023-00449-6. NPJ Biofilms Microbiomes. 2023. PMID: 37857690 Free PMC article.
-
The Kringle-like Domain Facilitates Post-endoplasmic Reticulum Changes to Premelanosome Protein (PMEL) Oligomerization and Disulfide Bond Configuration and Promotes Amyloid Formation.J Biol Chem. 2016 Feb 12;291(7):3595-612. doi: 10.1074/jbc.M115.692442. Epub 2015 Dec 22. J Biol Chem. 2016. PMID: 26694611 Free PMC article.
-
Cross-seeding between the functional amyloidogenic CRES and CRES3 family members and their regulation of Aβ assembly.J Biol Chem. 2021 Jan-Jun;296:100250. doi: 10.1074/jbc.RA120.015307. Epub 2021 Jan 9. J Biol Chem. 2021. PMID: 33384380 Free PMC article.
References
-
- Andersson EK, Bengtsson C, Evans ML, Chorell E, Sellstedt M, Lindgren AE, Hufnagel DA, Bhattacharya M, Tessier PM, Wittung-Stafshede P, Almqvist F, Chapman MR. Modulation of curli assembly and pellicle biofilm formation by chemical and protein chaperones. Chem Biol. 2013;20:1245–1254. - PMC - PubMed
-
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. - PubMed
-
- Broadley SA, Hartl FU. The role of molecular chaperones in human misfolding diseases. FEBS Lett. 2009;583:2647–2653. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

