DNA methylation profiles and biomarkers of oral squamous cell carcinoma
- PMID: 25612142
- PMCID: PMC4622594
- DOI: 10.1080/15592294.2015.1006506
DNA methylation profiles and biomarkers of oral squamous cell carcinoma
Abstract
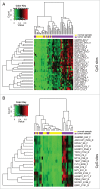
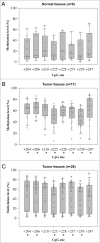
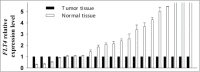
Oral squamous cell carcinoma (OSCC) constitutes >90% of oral cancers and is the sixth most common malignancy among males worldwide and the fourth leading cause of death due to cancer among males in Taiwan. However, most patients do not receive a diagnosis of OSCC until the late stages, which have a lower survival rate. The use of molecular marker analysis to identify early-stage OSCC would permit optimal timing for treatments and consequently prolong survival. The aim of this study was to identify biomarkers of OSCC using the Illumina GoldenGate Methylation Cancer Panel, which comprised a total of 1,505 CpG sites covering 807 genes. Samples of buccal mucosa resected from 40 OSCC patients and normal tissue samples obtained from 15 patients (normal mucosa from OSCC patients or from patients undergoing surgery unrelated to OSCC) were analyzed. Fms-related tyrosine kinase 4 (FLT4) methylation exhibited a perfect specificity for detecting OSCC, with an area under the receiver operating characteristic curve of 0.91 for both all-stage and early-stage OSCC. Methylation of 7 genes (ASCL1, FGF3, FLT4, GAS7, KDR, TERT, and TFPI2) constitutes the top-20 panels for detecting OSCC. The top-20 panels for detecting early-stage OSCC contain 8 genes: ADCYAP1, EPHA7, FLT4, GSTM2, KDR, MT1A, NPY, and TFPI2. FLT4 RNA expression and methylation level were validated using RT-PCR and a pyrosequencing methylation assay. The median level of FLT4 expression was 2.14-fold for normal relative to OSCC tissue samples (P < 0.0001). Among the 8 pyrosequenced FLT4 CpG sites, methylation level was much higher in the OSCC samples. In conclusion, methylation statuses of selected genes, and especially FLT4, KDR, and TFPI2, might be of great potential as biomarkers for early detection of buccal OSCC.
Keywords: DNA methylation array; FLT4; KDR; TFPI2; biomarkers; oral squamous cell carcinoma; pyrosequencing.
Figures
Similar articles
-
TRH site-specific methylation in oral and oropharyngeal squamous cell carcinoma.BMC Cancer. 2018 Aug 6;18(1):786. doi: 10.1186/s12885-018-4706-x. BMC Cancer. 2018. PMID: 30081853 Free PMC article.
-
CpG location and methylation level are crucial factors for the early detection of oral squamous cell carcinoma in brushing samples using bisulfite sequencing of a 13-gene panel.Clin Epigenetics. 2017 Aug 15;9:85. doi: 10.1186/s13148-017-0386-7. eCollection 2017. Clin Epigenetics. 2017. PMID: 28814981 Free PMC article.
-
[Comparative analysis of methylation profiles in tissues of oral leukoplakia and oral squamous cell carcinoma].Zhonghua Kou Qiang Yi Xue Za Zhi. 2018 Apr 9;53(4):248-253. doi: 10.3760/cma.j.issn.1002-0098.2018.04.007. Zhonghua Kou Qiang Yi Xue Za Zhi. 2018. PMID: 29690695 Chinese.
-
Prognostic biomarkers in oral squamous cell carcinoma: A systematic review.Oral Oncol. 2017 Sep;72:38-47. doi: 10.1016/j.oraloncology.2017.07.003. Epub 2017 Jul 12. Oral Oncol. 2017. PMID: 28797460 Review.
-
Hallmarks of Cancer-Related Newly Prognostic Factors of Oral Squamous Cell Carcinoma.Int J Mol Sci. 2018 Aug 16;19(8):2413. doi: 10.3390/ijms19082413. Int J Mol Sci. 2018. PMID: 30115834 Free PMC article. Review.
Cited by
-
Molecular pathways of oral cancer that predict prognosis and survival: A systematic review.J Carcinog. 2018 Dec 31;17:7. doi: 10.4103/jcar.JCar_17_18. eCollection 2018. J Carcinog. 2018. PMID: 30766450 Free PMC article. Review.
-
A transcriptome-based protein network that identifies new therapeutic targets in colorectal cancer.BMC Genomics. 2017 Sep 30;18(1):758. doi: 10.1186/s12864-017-4139-y. BMC Genomics. 2017. PMID: 28962550 Free PMC article.
-
GAS7 Deficiency Promotes Metastasis in MYCN-Driven Neuroblastoma.Cancer Res. 2021 Jun 1;81(11):2995-3007. doi: 10.1158/0008-5472.CAN-20-1890. Epub 2021 Feb 18. Cancer Res. 2021. PMID: 33602789 Free PMC article.
-
The REASON score: an epigenetic and clinicopathologic score to predict risk of poor survival in patients with early stage oral squamous cell carcinoma.Biomark Res. 2021 Jun 5;9(1):42. doi: 10.1186/s40364-021-00292-x. Biomark Res. 2021. PMID: 34090518 Free PMC article.
-
Interaction between tissue factor pathway inhibitor-2 gene polymorphisms and environmental factors associated with coronary atherosclerosis in a Chinese Han.J Thromb Thrombolysis. 2019 Jan;47(1):67-72. doi: 10.1007/s11239-018-1755-6. J Thromb Thrombolysis. 2019. PMID: 30343349
References
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136:E359-86; PMID:25220842; http://dx.doi.org/10.1002/ijc.29210 - DOI - PubMed
-
- Department of Health, Executive Yuan, R.O.C.(TAIWAN). 2009 Statistics of Causes of Death. Taipei, Taiwan, 2010.
-
- Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol 2009; 45:309-16; PMID:18804401; http://dx.doi.org/10.1016/j.oraloncology.2008.06.002 - DOI - PubMed
-
- Lo WL, Kao SY, Chi LY, Wong YK, Chang RC. Outcomes of oral squamous cell carcinoma in Taiwan after surgical therapy: factors affecting survival. Journal of Oral and Maxillofacial Surgery 2003; 61:751-8; PMID:12856245; http://dx.doi.org/10.1016/S0278-2391(03)00149-6 - DOI - PubMed
-
- Richards D. Patient delay in reporting oral cancer is poorly understood. Evid Based Dent 2007; 8:21; PMID:17380180; http://dx.doi.org/10.1038/sj.ebd.6400472 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
