Recoded organisms engineered to depend on synthetic amino acids
- PMID: 25607356
- PMCID: PMC4590768
- DOI: 10.1038/nature14095
Recoded organisms engineered to depend on synthetic amino acids
Erratum in
-
Corrigendum: Recoded organisms engineered to depend on synthetic amino acids.Nature. 2015 Nov 12;527(7577):264. doi: 10.1038/nature15537. Epub 2015 Sep 23. Nature. 2015. PMID: 26416756 No abstract available.
Abstract
Genetically modified organisms (GMOs) are increasingly used in research and industrial systems to produce high-value pharmaceuticals, fuels and chemicals. Genetic isolation and intrinsic biocontainment would provide essential biosafety measures to secure these closed systems and enable safe applications of GMOs in open systems, which include bioremediation and probiotics. Although safeguards have been designed to control cell growth by essential gene regulation, inducible toxin switches and engineered auxotrophies, these approaches are compromised by cross-feeding of essential metabolites, leaked expression of essential genes, or genetic mutations. Here we describe the construction of a series of genomically recoded organisms (GROs) whose growth is restricted by the expression of multiple essential genes that depend on exogenously supplied synthetic amino acids (sAAs). We introduced a Methanocaldococcus jannaschii tRNA:aminoacyl-tRNA synthetase pair into the chromosome of a GRO derived from Escherichia coli that lacks all TAG codons and release factor 1, endowing this organism with the orthogonal translational components to convert TAG into a dedicated sense codon for sAAs. Using multiplex automated genome engineering, we introduced in-frame TAG codons into 22 essential genes, linking their expression to the incorporation of synthetic phenylalanine-derived amino acids. Of the 60 sAA-dependent variants isolated, a notable strain harbouring three TAG codons in conserved functional residues of MurG, DnaA and SerS and containing targeted tRNA deletions maintained robust growth and exhibited undetectable escape frequencies upon culturing ∼10(11) cells on solid media for 7 days or in liquid media for 20 days. This is a significant improvement over existing biocontainment approaches. We constructed synthetic auxotrophs dependent on sAAs that were not rescued by cross-feeding in environmental growth assays. These auxotrophic GROs possess alternative genetic codes that impart genetic isolation by impeding horizontal gene transfer and now depend on the use of synthetic biochemical building blocks, advancing orthogonal barriers between engineered organisms and the environment.
Figures
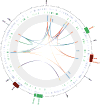



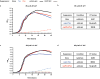
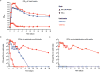
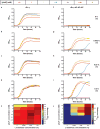
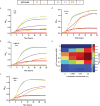


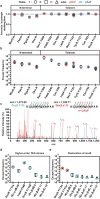


Comment in
-
Synthetic biology: GMOs in lockdown.Nat Rev Microbiol. 2015 Mar;13(3):125. doi: 10.1038/nrmicro3443. Epub 2015 Feb 9. Nat Rev Microbiol. 2015. PMID: 25659321 No abstract available.
-
Synthetic biology: GMOs in lockdown.Nat Rev Genet. 2015 Mar;16(3):127. doi: 10.1038/nrg3909. Nat Rev Genet. 2015. PMID: 25690387 No abstract available.
Similar articles
-
Biocontainment of genetically modified organisms by synthetic protein design.Nature. 2015 Feb 5;518(7537):55-60. doi: 10.1038/nature14121. Epub 2015 Jan 21. Nature. 2015. PMID: 25607366 Free PMC article.
-
Synthetic biology: GMOs in lockdown.Nat Rev Genet. 2015 Mar;16(3):127. doi: 10.1038/nrg3909. Nat Rev Genet. 2015. PMID: 25690387 No abstract available.
-
Synthetic biology: GMOs in lockdown.Nat Rev Microbiol. 2015 Mar;13(3):125. doi: 10.1038/nrmicro3443. Epub 2015 Feb 9. Nat Rev Microbiol. 2015. PMID: 25659321 No abstract available.
-
Engineering the Genetic Code in Cells and Animals: Biological Considerations and Impacts.Acc Chem Res. 2017 Nov 21;50(11):2767-2775. doi: 10.1021/acs.accounts.7b00376. Epub 2017 Oct 6. Acc Chem Res. 2017. PMID: 28984438 Free PMC article. Review.
-
Recent advances in genetic code engineering in Escherichia coli.Curr Opin Biotechnol. 2012 Oct;23(5):751-7. doi: 10.1016/j.copbio.2011.12.027. Epub 2012 Jan 9. Curr Opin Biotechnol. 2012. PMID: 22237016 Review.
Cited by
-
Selection platforms for directed evolution in synthetic biology.Biochem Soc Trans. 2016 Aug 15;44(4):1165-75. doi: 10.1042/BST20160076. Biochem Soc Trans. 2016. PMID: 27528765 Free PMC article. Review.
-
Towards Biocontained Cell Factories: An Evolutionarily Adapted Escherichia coli Strain Produces a New-to-nature Bioactive Lantibiotic Containing Thienopyrrole-Alanine.Sci Rep. 2016 Sep 16;6:33447. doi: 10.1038/srep33447. Sci Rep. 2016. PMID: 27634138 Free PMC article.
-
Yeast as a cell factory: current state and perspectives.Microb Cell Fact. 2015 Jun 30;14:94. doi: 10.1186/s12934-015-0281-x. Microb Cell Fact. 2015. PMID: 26122609 Free PMC article. Review.
-
Genetic Code Expansion Through Quadruplet Codon Decoding.J Mol Biol. 2022 Apr 30;434(8):167346. doi: 10.1016/j.jmb.2021.167346. Epub 2021 Nov 8. J Mol Biol. 2022. PMID: 34762896 Free PMC article. Review.
-
Genetic-code-expanded cell-based therapy for treating diabetes in mice.Nat Chem Biol. 2022 Jan;18(1):47-55. doi: 10.1038/s41589-021-00899-z. Epub 2021 Nov 15. Nat Chem Biol. 2022. PMID: 34782743
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

