Prp40 pre-mRNA processing factor 40 homolog B (PRPF40B) associates with SF1 and U2AF65 and modulates alternative pre-mRNA splicing in vivo
- PMID: 25605964
- PMCID: PMC4338339
- DOI: 10.1261/rna.047258.114
Prp40 pre-mRNA processing factor 40 homolog B (PRPF40B) associates with SF1 and U2AF65 and modulates alternative pre-mRNA splicing in vivo
Abstract
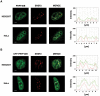
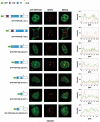
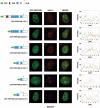
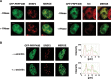
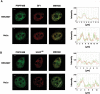
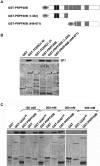
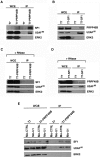
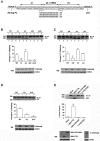
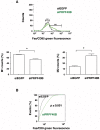
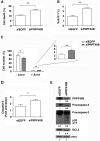
The first stable complex formed during the assembly of spliceosomes onto pre-mRNA substrates in mammals includes U1 snRNP, which recognizes the 5' splice site, and the splicing factors SF1 and U2AF, which bind the branch point sequence, polypyrimidine tract, and 3' splice site. The 5' and 3' splice site complexes are thought to be joined together by protein-protein interactions mediated by factors that ensure the fidelity of the initial splice site recognition. In this study, we identified and characterized PRPF40B, a putative mammalian ortholog of the U1 snRNP-associated yeast splicing factor Prp40. PRPF40B is highly enriched in speckles with a behavior similar to splicing factors. We demonstrated that PRPF40B interacts directly with SF1 and associates with U2AF(65). Accordingly, PRPF40B colocalizes with these splicing factors in the cell nucleus. Splicing assays with reporter minigenes revealed that PRPF40B modulates alternative splice site selection. In the case of Fas regulation of alternative splicing, weak 5' and 3' splice sites and exonic sequences are required for PRPF40B function. Placing our data in a functional context, we also show that PRPF40B depletion increased Fas/CD95 receptor number and cell apoptosis, which suggests the ability of PRPF40B to alter the alternative splicing of key apoptotic genes to regulate cell survival.
Keywords: PRPF40B; alternative splicing; mRNA processing.
© 2015 Becerra et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

Similar articles
-
Splicing inhibition of U2AF65 leads to alternative exon skipping.Proc Natl Acad Sci U S A. 2015 Aug 11;112(32):9926-31. doi: 10.1073/pnas.1500639112. Epub 2015 Jul 27. Proc Natl Acad Sci U S A. 2015. PMID: 26216990 Free PMC article.
-
Characterization of a U2AF-independent commitment complex (E') in the mammalian spliceosome assembly pathway.Mol Cell Biol. 2005 Jan;25(1):233-40. doi: 10.1128/MCB.25.1.233-240.2005. Mol Cell Biol. 2005. PMID: 15601845 Free PMC article.
-
Mammalian splicing factor SF1 interacts with SURP domains of U2 snRNP-associated proteins.Nucleic Acids Res. 2015 Dec 2;43(21):10456-73. doi: 10.1093/nar/gkv952. Epub 2015 Sep 29. Nucleic Acids Res. 2015. PMID: 26420826 Free PMC article.
-
RNA-protein interactions that regulate pre-mRNA splicing.Gene Expr. 2002;10(1-2):79-92. Gene Expr. 2002. PMID: 11868989 Free PMC article. Review.
-
Regulation of alternative mRNA splicing: old players and new perspectives.FEBS Lett. 2018 Sep;592(17):2987-3006. doi: 10.1002/1873-3468.13119. Epub 2018 Jun 16. FEBS Lett. 2018. PMID: 29856907 Review.
Cited by
-
Active 5' splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes.Nucleic Acids Res. 2017 Mar 17;45(5):2757-2775. doi: 10.1093/nar/gkw895. Nucleic Acids Res. 2017. PMID: 27907902 Free PMC article.
-
Light in the transcription landscape: chromatin, RNA polymerase II and splicing throughout Arabidopsis thaliana's life cycle.Transcription. 2020 Jun-Aug;11(3-4):117-133. doi: 10.1080/21541264.2020.1796473. Epub 2020 Aug 4. Transcription. 2020. PMID: 32748694 Free PMC article. Review.
-
PRPF40A induces inclusion of exons in GC-rich regions important for human myeloid cell differentiation.Nucleic Acids Res. 2024 Aug 27;52(15):8800-8814. doi: 10.1093/nar/gkae557. Nucleic Acids Res. 2024. PMID: 38943321 Free PMC article.
-
CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing.Nat Commun. 2017 Oct 19;8(1):1035. doi: 10.1038/s41467-017-01241-9. Nat Commun. 2017. PMID: 29051543 Free PMC article.
-
Cholesteryl oleate-loaded cationic solid lipid nanoparticles as carriers for efficient gene-silencing therapy.Int J Nanomedicine. 2018 May 30;13:3223-3233. doi: 10.2147/IJN.S158884. eCollection 2018. Int J Nanomedicine. 2018. PMID: 29881274 Free PMC article.
References
-
- Abovich N, Rosbash M 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89: 403–412. - PubMed
-
- Boise LH, González-García M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nuñez G, Thompson CB 1993. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74: 597–608. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
